Genome wide scan in a Flemish inflammatory bowel disease population: support for the IBD4 locus, population heterogeneity, and epistasis
- PMID: 15194648
- PMCID: PMC1774099
- DOI: 10.1136/gut.2003.034033
Genome wide scan in a Flemish inflammatory bowel disease population: support for the IBD4 locus, population heterogeneity, and epistasis
Abstract
Background and aims: Genome wide scans in inflammatory bowel disease (IBD) have indicated various susceptibility regions with replication of 16cen (IBD1), 12q (IBD2), 6p (IBD3), 14q11 (IBD4), and 3p21. As no linkage was previously found on IBD regions 3, 7, 12, and 16 in Flemish IBD families, a genome wide scan was performed to detect other susceptibility regions in this population.
Methods: A cohort of 149 IBD affected relative pairs, all recruited from the Northern Flemish part of Belgium, were genotyped using microsatellite markers at 12 cM intervals, and analysed by Genehunter non-parametric linkage software. All families were further genotyped for the three main Crohn's disease associated variants in the NOD2/CARD15 gene.
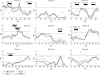
Results: Nominal evidence for linkage was observed on chromosomes 1 (D1S197: multipoint non-parametric linkage (NPL) score 2.57, p = 0.004; and at D1S305-D1S252: NPL 2.97, p = 0.001), 4q (D4S406: NPL 1.95, p = 0.03), 6q16 (D6S314: NPL 2.44, p = 0.007), 10p12 (D10S197: NPL 2.05, p = 0.02), 11q22 (D11S35-D11S927: NPL 1.95, p = 0.02) 14q11-12 (D14S80: NPL 2.41, p = 0.008), 20p12 (D20S192: NPL 2.7, p = 0.003), and Xq (DXS990: NPL 1.70, p = 0.04). A total of 51.4% of patients carried at least one NOD2/CARD15 variant. Furthermore, epistasis was observed between susceptibility regions 6q/10p and 20p/10p.
Conclusion: Genome scanning in a Flemish IBD population found nominal evidence for linkage on 1p, 4q, 10p12, and 14q11, overlapping with other genome scan results, with linkage on 14q11-12 supporting the IBD4 locus. The results further show that epistasis is contributing to the complex model of IBD and indicate that population heterogeneity is not to be underestimated. Finally, NOD2/CARD15 is clearly implicated in the Flemish IBD population.
Figures


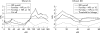
Similar articles
-
High-density genome scan in Crohn disease shows confirmed linkage to chromosome 14q11-12.Am J Hum Genet. 2000 Jun;66(6):1857-62. doi: 10.1086/302947. Epub 2000 Apr 3. Am J Hum Genet. 2000. PMID: 10747815 Free PMC article.
-
The IBD international genetics consortium provides further evidence for linkage to IBD4 and shows gene-environment interaction.Inflamm Bowel Dis. 2005 Jan;11(1):1-7. doi: 10.1097/00054725-200501000-00001. Inflamm Bowel Dis. 2005. PMID: 15674107
-
Identification of genetic loci for basal cell nevus syndrome and inflammatory bowel disease in a single large pedigree.Hum Genet. 2006 Aug;120(1):31-41. doi: 10.1007/s00439-006-0163-8. Epub 2006 May 30. Hum Genet. 2006. PMID: 16733713
-
Inflammatory bowel disease gene hunting by linkage analysis: rationale, methodology, and present status of the field.Inflamm Bowel Dis. 2004 May;10(3):300-11. doi: 10.1097/00054725-200405000-00019. Inflamm Bowel Dis. 2004. PMID: 15290927 Review.
-
[Genetics of inflammatory bowel disease].Tunis Med. 2004 Jul;82(7):635-41. Tunis Med. 2004. PMID: 15552020 Review. French.
Cited by
-
Genetic and Epigenetic Etiology of Inflammatory Bowel Disease: An Update.Genes (Basel). 2022 Dec 16;13(12):2388. doi: 10.3390/genes13122388. Genes (Basel). 2022. PMID: 36553655 Free PMC article. Review.
-
Interpretable network-guided epistasis detection.Gigascience. 2022 Feb 4;11:giab093. doi: 10.1093/gigascience/giab093. Gigascience. 2022. PMID: 35134928 Free PMC article.
-
Intestinal Epithelial Cell Endoplasmic Reticulum Stress and Inflammatory Bowel Disease Pathogenesis: An Update Review.Front Immunol. 2017 Oct 25;8:1271. doi: 10.3389/fimmu.2017.01271. eCollection 2017. Front Immunol. 2017. PMID: 29118753 Free PMC article. Review.
-
Genetic update on inflammatory factors in ulcerative colitis: Review of the current literature.World J Gastrointest Pathophysiol. 2014 Aug 15;5(3):304-21. doi: 10.4291/wjgp.v5.i3.304. World J Gastrointest Pathophysiol. 2014. PMID: 25133031 Free PMC article. Review.
-
Endoplasmic reticulum stress, unfolded protein response and altered T cell differentiation in necrotizing enterocolitis.PLoS One. 2013 Oct 23;8(10):e78491. doi: 10.1371/journal.pone.0078491. eCollection 2013. PLoS One. 2013. PMID: 24194940 Free PMC article.
References
-
- Calkins BM, Mendelhoff AI. The epidemiology of ideopathic inflammatory bowel disease. In: Kirsner JB, Shorter RG, eds. Inflammatory bowel disease. 4th ed. Baltimore: Williams & Wilkins, 1995:31–68.
-
- Binder V . Genetic epidemiology in inflammatory bowel disease. Dig Dis 1998;16:351–5. - PubMed
-
- Vermeire S , Peeters M, Vlietinck R, et al. Anti-Saccharomyces cerevisiae antibodies (ASCA), phenotypes of IBD and intestinal permeability: a study in familial IBD. Inflamm Bowel Dis 2001;7:8–15. - PubMed
-
- Duerr RH, Targan SR, Landers CJ, et al. Anti-neutrophil cytoplasmic antibodies in ulcerative colitis: comparison with other colitides/diarrhoeal illnesses. Gastroenterology 1992;100:1590–6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

