A bioinformatics analysis of memory consolidation reveals involvement of the transcription factor c-rel
- PMID: 15102909
- PMCID: PMC6729420
- DOI: 10.1523/JNEUROSCI.5646-03.2004
A bioinformatics analysis of memory consolidation reveals involvement of the transcription factor c-rel
Abstract
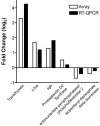
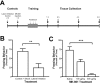
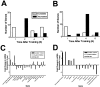
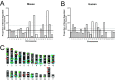
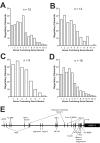
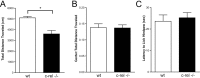
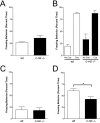
Consolidation of long-term memory (LTM) is a complex process requiring synthesis of new mRNAs and proteins. Many studies have characterized the requirement for de novo mRNA and protein synthesis; however, few studies have comprehensively identified genes regulated during LTM consolidation. We show that consolidation of long-term contextual memory in the hippocampus triggers altered expression of numerous genes encompassing many aspects of neuronal function. Like contextual memory formation, this altered gene expression required NMDA receptor activation and was specific for situations in which the animal formed an association between a physical context and a sensory stimulus. Using a bioinformatics approach, we found that regulatory elements for several transcription factors are over-represented in the upstream region of genes regulated during consolidation of LTM. Using a knock-out mouse, we found that c-rel, one of the transcription factors identified in our bioinformatics study, is necessary for hippocampus-dependent long-term memory formation.
Figures
Similar articles
-
c-Rel, an NF-kappaB family transcription factor, is required for hippocampal long-term synaptic plasticity and memory formation.Learn Mem. 2008 Jul 11;15(7):539-49. doi: 10.1101/lm.866408. Print 2008 Jul. Learn Mem. 2008. PMID: 18626097 Free PMC article.
-
N-glycosylation in the hippocampus is required for the consolidation and reconsolidation of contextual fear memory.Neurobiol Learn Mem. 2016 Nov;135:57-65. doi: 10.1016/j.nlm.2016.06.018. Epub 2016 Jun 21. Neurobiol Learn Mem. 2016. PMID: 27343988
-
Consolidation of CS and US representations in associative fear conditioning.Hippocampus. 2004;14(5):557-69. doi: 10.1002/hipo.10208. Hippocampus. 2004. PMID: 15301434
-
Gene expression profiles--a new dynamic and functional dimension to the exploration of learning and memory.Rev Neurosci. 2002;13(3):209-19. doi: 10.1515/revneuro.2002.13.3.209. Rev Neurosci. 2002. PMID: 12405225 Review.
-
Memory formation and the regulation of gene expression.Cell Mol Life Sci. 1999 Apr;55(4):575-92. doi: 10.1007/s000180050316. Cell Mol Life Sci. 1999. PMID: 10357228 Free PMC article. Review.
Cited by
-
Gene Expression of Neurogenesis Related to Exercise Intensity in a Cerebral Infarction Rat Model.Int J Mol Sci. 2024 Aug 19;25(16):8997. doi: 10.3390/ijms25168997. Int J Mol Sci. 2024. PMID: 39201683 Free PMC article.
-
Potential therapeutic effects of peroxisome proliferator-activated receptors on corneal diseases.Exp Biol Med (Maywood). 2024 Jun 27;249:10142. doi: 10.3389/ebm.2024.10142. eCollection 2024. Exp Biol Med (Maywood). 2024. PMID: 38993197 Free PMC article. Review.
-
The Effect of Simvastatin on the Dynamics of NF-κB-Regulated Neurodegenerative and Neuroprotective Processes in the Acute Phase of Ischemic Stroke.Mol Neurobiol. 2023 Sep;60(9):4935-4951. doi: 10.1007/s12035-023-03371-2. Epub 2023 May 19. Mol Neurobiol. 2023. PMID: 37204689 Free PMC article.
-
Translatome analysis reveals microglia and astrocytes to be distinct regulators of inflammation in the hyperacute and acute phases after stroke.Glia. 2023 Aug;71(8):1960-1984. doi: 10.1002/glia.24377. Epub 2023 Apr 17. Glia. 2023. PMID: 37067534 Free PMC article.
-
Oral Peroxisome Proliferator-Activated Receptor-α Agonist Enhances Corneal Nerve Regeneration in Patients With Type 2 Diabetes.Diabetes. 2023 Jul 1;72(7):932-946. doi: 10.2337/db22-0611. Diabetes. 2023. PMID: 36445944 Free PMC article.
References
-
- Abel T, Kandel E (1998) Positive and negative regulatory mechanisms that mediate long-term memory storage. Brain Res Brain Res Rev 26: 360-378. - PubMed
-
- Abraham WC, Dragunow M, Tate WP (1991) The role of immediate early genes in the stabilization of long-term potentiation. Mol Neurobiol 5: 297-314. - PubMed
-
- Accetta SG, Abeche AM, Buchabqui JA, Hammes L, Pratti R, Afler T, Capp E (2002) Memory loss and ataxia after hyperemesis gravidarum: a case of Wernicke-Korsakoff syndrome. Eur J Obstet Gynecol Reprod Biol 102: 100-101. - PubMed
-
- Adams JP, Sweatt JD (2002) Molecular psychology: roles for the ERK MAP kinase cascade in memory. Annu Rev Pharmacol Toxicol 42: 135-163. - PubMed
-
- Alberini CM, Ghirardi M, Metz R, Kandel ER (1994) C/EBP is an immediate-early gene required for the consolidation of long-term facilitation in Aplysia Cell 76: 1099-1114. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

