Cultivation and growth characteristics of a diverse group of oligotrophic marine Gammaproteobacteria
- PMID: 14711672
- PMCID: PMC321273
- DOI: 10.1128/AEM.70.1.432-440.2004
Cultivation and growth characteristics of a diverse group of oligotrophic marine Gammaproteobacteria
Abstract
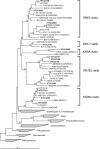
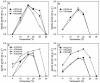
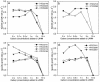
Forty-four novel strains of Gammaproteobacteria were cultivated from coastal and pelagic regions of the Pacific Ocean using high-throughput culturing methods that rely on dilution to extinction in very low nutrient media. Phylogenetic analysis showed that the isolates fell into five rRNA clades, all of which contained rRNA gene sequences reported previously from seawater environmental gene clone libraries (SAR92, OM60, OM182, BD1-7, and KI89A). Bootstrap analyses of phylogenetic reliability did not support collapsing these five clades into a single clade, and they were therefore named the oligotrophic marine Gammaproteobacteria (OMG) group. Twelve cultures chosen to represent the five clades were successively purified in liquid culture, and their growth characteristics were determined at different temperatures and dissolved organic carbon concentrations. The isolates in the OMG group were physiologically diverse heterotrophs, and their physiological properties generally followed their phylogenetic relationships. None of the isolates in the OMG group formed colonies on low- or high-nutrient agar upon their first isolation from seawater, while 7 of 12 isolates that were propagated for laboratory testing eventually produced colonies on 1/10 R2A agar. The isolates grew relatively slowly in natural seawater media (1.23 to 2.63 day(-1)), and none of them grew in high-nutrient media (>351 mg of C liter(-1)). The isolates were psychro- to mesophilic and obligately oligotrophic; many of them were of ultramicrobial size (<0.1 micro m(3)). This cultivation study revealed that sporadically detected Gammaproteobacteria gene clones from seawater are part of a phylogenetically diverse constellation of organisms mainly composed of oligotrophic and ultramicrobial lineages that are culturable under specific cultivation conditions.
Figures

Similar articles
-
Expansion of Cultured Bacterial Diversity by Large-Scale Dilution-to-Extinction Culturing from a Single Seawater Sample.Microb Ecol. 2016 Jan;71(1):29-43. doi: 10.1007/s00248-015-0695-3. Epub 2015 Nov 14. Microb Ecol. 2016. PMID: 26573832
-
The phylogenetic and ecological context of cultured and whole genome-sequenced planktonic bacteria from the coastal NW Mediterranean Sea.Syst Appl Microbiol. 2014 May;37(3):216-28. doi: 10.1016/j.syapm.2013.11.005. Epub 2014 Jan 23. Syst Appl Microbiol. 2014. PMID: 24462268
-
Biogeography and phylogeny of the NOR5/OM60 clade of Gammaproteobacteria.Syst Appl Microbiol. 2009 Apr;32(2):124-39. doi: 10.1016/j.syapm.2008.12.001. Syst Appl Microbiol. 2009. PMID: 19216045
-
High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates.Appl Environ Microbiol. 2002 Aug;68(8):3878-85. doi: 10.1128/AEM.68.8.3878-3885.2002. Appl Environ Microbiol. 2002. PMID: 12147485 Free PMC article.
-
Culturing marine bacteria - an essential prerequisite for biodiscovery.Microb Biotechnol. 2010 Sep;3(5):564-75. doi: 10.1111/j.1751-7915.2010.00188.x. Epub 2010 Jun 21. Microb Biotechnol. 2010. PMID: 21255353 Free PMC article. Review.
Cited by
-
Microbial Community Structure in the Taklimakan Desert: The Importance of Nutrient Levels in Medium and Culture Methods.Biology (Basel). 2024 Oct 6;13(10):797. doi: 10.3390/biology13100797. Biology (Basel). 2024. PMID: 39452106 Free PMC article.
-
Diversity and distribution of a prevalent Microviridae group across the global oceans.Commun Biol. 2024 Oct 23;7(1):1377. doi: 10.1038/s42003-024-07085-6. Commun Biol. 2024. PMID: 39443614 Free PMC article.
-
Congregibacter variabilis sp. nov. and Congregibacter brevis sp. nov. Within the OM60/NOR5 Clade, Isolated from Seawater, and Emended Description of the Genus Congregibacter.J Microbiol. 2024 Sep;62(9):739-748. doi: 10.1007/s12275-024-00158-5. Epub 2024 Jul 18. J Microbiol. 2024. PMID: 39023694
-
Genome sequences of the first Autographiviridae phages infecting marine Roseobacter.Microb Genom. 2024 Apr;10(4):001240. doi: 10.1099/mgen.0.001240. Microb Genom. 2024. PMID: 38630615 Free PMC article.
-
Comparative genomics of the genus Halioglobus reveals the genetic basis for the reclassification of Halioglobus pacificus as Parahalioglobus pacificus gen. nov. comb. nov.Int Microbiol. 2024 Apr 1. doi: 10.1007/s10123-024-00516-8. Online ahead of print. Int Microbiol. 2024. PMID: 38558270
References
-
- Béjà, O., M. T. Suzuki, E. V. Koonin, L. Aravind, A. Hadd, L. P. Nguyen, R. Villacorta, M. Amjadi, C. Garrigues, S. B. Jovanovich, R. A. Feldman, and E. F. DeLong. 2000. Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol. 2:516-529. - PubMed
-
- Bowman, J. P., S. M. Rea, S. A. McCammon, and T. A. McMeekin. 2000. Diversity and community structure within anoxic sediment from marine salinity meromictic lakes and a coastal meromictic marine basin, Vestfold Hilds, Eastern Antarctica. Environ. Microbiol. 2:227-237. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases


