Dissection of genomewide-scan data in extended families reveals a major locus and oligogenic susceptibility for age-related macular degeneration
- PMID: 14691731
- PMCID: PMC1181910
- DOI: 10.1086/380912
Dissection of genomewide-scan data in extended families reveals a major locus and oligogenic susceptibility for age-related macular degeneration
Abstract
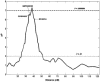
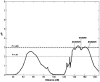
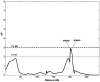
To examine the genetic basis of age-related macular degeneration (ARMD), a degenerative disease of the retinal pigment epithelium and neurosensory retina, we conducted a genomewide scan in 34 extended families (297 individuals, 349 sib pairs) ascertained through index cases with neovascular disease or geographic atrophy. Family and medical history was obtained from index cases and family members. Fundus photographs were taken of all participating family members, and these were graded for severity by use of a quantitative scale. Model-free linkage analysis was performed, and tests of heterogeneity and epistasis were conducted. We have evidence of a major locus on chromosome 15q (GATA50C03 multipoint P=1.98x10-7; empirical P< or =1.0x10-5; single-point P=3.6x10-7). This locus was present as a weak linkage signal in our previous genome scan for ARMD, in the Beaver Dam Eye Study sample (D15S659, multipoint P=.047), but is otherwise novel. In this genome scan, we observed a total of 13 regions on 11 chromosomes (1q31, 2p21, 4p16, 5q34, 9p24, 9q31, 10q26, 12q13, 12q23, 15q21, 16p12, 18p11, and 20q13), with a nominal multipoint significance level of P< or =.01 or LOD > or =1.18. Family-by-family analysis of the data, performed using model-free linkage methods, suggests that there is evidence of heterogeneity in these families. For example, a single family (family 460) individually shows linkage evidence at 8 loci, at the level of P<.0001. We conducted tests for heterogeneity, which suggest that ARMD susceptibility loci on chromosomes 9p24, 10q26, and 15q21 are not present in all families. We tested for mutations in linked families and examined SNPs in two candidate genes, hemicentin-1 and EFEMP1, in subsamples (145 and 189 sib pairs, respectively) of the data. Mutations were not observed in any of the 11 exons of EFEMP1 nor in exon 104 of hemicentin-1. The SNP analysis for hemicentin-1 on 1q31 suggests that variants within or in very close proximity to this gene cause ARMD pathogenesis. In summary, we have evidence for a major ARMD locus on 15q21, which, coupled with numerous other loci segregating in these families, suggests complex oligogenic patterns of inheritance for ARMD.
Figures


Similar articles
-
Age-related maculopathy: an expanded genome-wide scan with evidence of susceptibility loci within the 1q31 and 17q25 regions.Am J Ophthalmol. 2001 Nov;132(5):682-92. doi: 10.1016/s0002-9394(01)01214-4. Am J Ophthalmol. 2001. PMID: 11704029
-
Ordered subset linkage analysis supports a susceptibility locus for age-related macular degeneration on chromosome 16p12.BMC Genet. 2004 Jul 6;5:18. doi: 10.1186/1471-2156-5-18. BMC Genet. 2004. PMID: 15238159 Free PMC article.
-
Age-related maculopathy: a genomewide scan with continued evidence of susceptibility loci within the 1q31, 10q26, and 17q25 regions.Am J Hum Genet. 2004 Aug;75(2):174-89. doi: 10.1086/422476. Epub 2004 May 27. Am J Hum Genet. 2004. PMID: 15168325 Free PMC article.
-
HEMICENTIN-1 (FIBULIN-6) and the 1q31 AMD locus in the context of complex disease: review and perspective.Ophthalmic Genet. 2005 Jun;26(2):101-5. doi: 10.1080/13816810590968023. Ophthalmic Genet. 2005. PMID: 16020313 Review.
-
The genetics of inherited macular dystrophies.J Med Genet. 2003 Sep;40(9):641-50. doi: 10.1136/jmg.40.9.641. J Med Genet. 2003. PMID: 12960208 Free PMC article. Review.
Cited by
-
The Impact of ARMS2 (rs10490924), VEGFA (rs3024997), TNFRSF1B (rs1061622), TNFRSF1A (rs4149576), and IL1B1 (rs1143623) Polymorphisms and Serum Levels on Age-Related Macular Degeneration Development and Therapeutic Responses.Int J Mol Sci. 2024 Sep 9;25(17):9750. doi: 10.3390/ijms25179750. Int J Mol Sci. 2024. PMID: 39273697 Free PMC article.
-
CFH (rs1061170, rs1410996), KDR (rs2071559, rs1870377) and KDR and CFH Serum Levels in AMD Development and Treatment Efficacy.Biomedicines. 2024 Apr 24;12(5):948. doi: 10.3390/biomedicines12050948. Biomedicines. 2024. PMID: 38790910 Free PMC article.
-
Bruch's Membrane: A Key Consideration with Complement-Based Therapies for Age-Related Macular Degeneration.J Clin Med. 2023 Apr 14;12(8):2870. doi: 10.3390/jcm12082870. J Clin Med. 2023. PMID: 37109207 Free PMC article. Review.
-
Molecular Genetic Mechanisms in Age-Related Macular Degeneration.Genes (Basel). 2022 Jul 12;13(7):1233. doi: 10.3390/genes13071233. Genes (Basel). 2022. PMID: 35886016 Free PMC article. Review.
-
Complement System and Potential Therapeutics in Age-Related Macular Degeneration.Int J Mol Sci. 2021 Jun 25;22(13):6851. doi: 10.3390/ijms22136851. Int J Mol Sci. 2021. PMID: 34202223 Free PMC article. Review.
References
Electronic-Database Information
-
- Ensembl Genome Browser, http://www.ensembl.org/ (hemicentin-1 transcript ID ENSG00000143341)
-
- Marshfield Center for Medical Genetics, http://research.marshfieldclinic.org/genetics/
-
- Online Mendelian Inheritance of Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for EFEMP1)
References
-
- Allikmets R, Singh N, Sun H, Shroyer NF, Hutchinson A, Chidambaram A, Gerrard B, Baird L, Stauffer D, Peiffer A, Rattner A, Smallwood P, Li Y, Anderson KL, Lewis RA, Nathans J, Leppert M, Dean M, Lupski JR (1997b) A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat Genet 15:236–246 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous


