Proteorhodopsin genes are distributed among divergent marine bacterial taxa
- PMID: 14566056
- PMCID: PMC240704
- DOI: 10.1073/pnas.2133554100
Proteorhodopsin genes are distributed among divergent marine bacterial taxa
Abstract
Proteorhodopsin (PR) is a retinal-binding bacterial integral membrane protein that functions as a light-driven proton pump. The gene encoding this photoprotein was originally discovered on a large genome fragment derived from an uncultured marine gamma-proteobacterium of the SAR86 group. Subsequently, many variants of the PR gene have been detected in marine plankton, via PCR-based gene surveys. It has not been clear, however, whether these different PR genes are widely distributed among different bacterial groups, or whether they have a restricted taxonomic distribution. We report here comparative analyses of PR-bearing genomic fragments recovered directly from planktonic bacteria inhabiting the California coast, the central Pacific Ocean, and waters offshore the Antarctica Peninsula. Sequence analysis of an Antarctic genome fragment harboring PR (ANT32C12) revealed moderate conservation in gene order and identity, compared with a previously reported PR-containing genome fragment from a Monterey Bay gamma-proteobacterium (EBAC31A08). Outside the limited region of synteny shared between these clones, however, no significant DNA or protein identity was evident. Analysis of a third PR-containing genome fragment (HOT2C01) from the North Pacific subtropical gyre showed even more divergence from the gamma-proteobacterial PR-flanking region. Subsequent phylogenetic and comparative genomic analyses revealed that the Central North Pacific PR-containing genome fragment (HOT2C01) originated from a planktonic alpha-proteobacterium. These data indicate that PR genes are distributed among a variety of divergent marine bacterial taxa, including both alpha- and gamma-proteobacteria. Our analyses also demonstrate the utility of cultivation-independent comparative genomic approaches for assessing gene content and distribution in naturally occurring microbes.
Figures
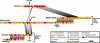
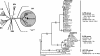

Similar articles
-
Different SAR86 subgroups harbour divergent proteorhodopsins.Environ Microbiol. 2004 Sep;6(9):903-10. doi: 10.1111/j.1462-2920.2004.00676.x. Environ Microbiol. 2004. PMID: 15305915
-
Abundant proteorhodopsin genes in the North Atlantic Ocean.Environ Microbiol. 2008 Jan;10(1):99-109. doi: 10.1111/j.1462-2920.2007.01436.x. Environ Microbiol. 2008. PMID: 18211270
-
Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea.Nature. 2006 Feb 16;439(7078):847-50. doi: 10.1038/nature04435. Nature. 2006. PMID: 16482157
-
Phylogenetic analyses of ribosomal DNA-containing bacterioplankton genome fragments from a 4000 m vertical profile in the North Pacific Subtropical Gyre.Environ Microbiol. 2008 Sep;10(9):2313-30. doi: 10.1111/j.1462-2920.2008.01657.x. Epub 2008 May 20. Environ Microbiol. 2008. PMID: 18494796
-
Marine Bacterial and Archaeal Ion-Pumping Rhodopsins: Genetic Diversity, Physiology, and Ecology.Microbiol Mol Biol Rev. 2016 Sep 14;80(4):929-54. doi: 10.1128/MMBR.00003-16. Print 2016 Dec. Microbiol Mol Biol Rev. 2016. PMID: 27630250 Free PMC article. Review.
Cited by
-
Proton-pumping rhodopsins promote the growth and survival of phytoplankton in a highly variable ocean.ISME J. 2024 Jan 8;18(1):wrae079. doi: 10.1093/ismejo/wrae079. ISME J. 2024. PMID: 38696358 Free PMC article. No abstract available.
-
A timeline of bacterial and archaeal diversification in the ocean.Elife. 2023 Dec 7;12:RP88268. doi: 10.7554/eLife.88268. Elife. 2023. PMID: 38059790 Free PMC article.
-
Widespread use of proton-pumping rhodopsin in Antarctic phytoplankton.Proc Natl Acad Sci U S A. 2023 Sep 26;120(39):e2307638120. doi: 10.1073/pnas.2307638120. Epub 2023 Sep 18. Proc Natl Acad Sci U S A. 2023. PMID: 37722052 Free PMC article.
-
Carotenoid binding in Gloeobacteria rhodopsin provides insights into divergent evolution of xanthorhodopsin types.Commun Biol. 2022 May 30;5(1):512. doi: 10.1038/s42003-022-03429-2. Commun Biol. 2022. PMID: 35637261 Free PMC article.
-
Proteorhodopsin Phototrophy in Antarctic Coastal Waters.mSphere. 2021 Aug 25;6(4):e0052521. doi: 10.1128/mSphere.00525-21. Epub 2021 Aug 18. mSphere. 2021. PMID: 34406852 Free PMC article.
References
-
- Spudich, J. L., Yang, C. S., Jung, K. H. & Spudich, E. N. (2000) Annu. Rev. Cell Dev. Biol. 16 365–392. - PubMed
-
- Béjà, O., Aravind, L., Koonin, E. V., Suzuki, M. T., Hadd, A., Nguyen, L. P., Jovanovich, S. B., Gates, C. M., Feldman, R. A., Spudich, J. L., et al. (2000) Science 289 1902–1906. - PubMed
-
- Béjà, O., Spudich, E. N., Spudich, J. L., Leclerc, M. & DeLong, E. F. (2001) Nature 411 786–789. - PubMed
-
- Sabehi, G., Massana, R., Bielawski, J. P., Rosenberg, M., DeLong, E. D. & Béjà, O. (2003) Environ Microbiol. 5 842–849. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials


