Prevalence of pandemic thermostable direct hemolysin-producing Vibrio parahaemolyticus O3:K6 in seafood and the coastal environment in Japan
- PMID: 12839757
- PMCID: PMC165169
- DOI: 10.1128/AEM.69.7.3883-3891.2003
Prevalence of pandemic thermostable direct hemolysin-producing Vibrio parahaemolyticus O3:K6 in seafood and the coastal environment in Japan
Abstract
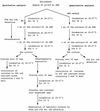
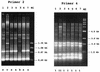
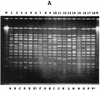
Although thermostable direct hemolysin (TDH)-producing Vibrio parahaemolyticus has caused many infections in Asian countries, the United States, and other countries, it has been difficult to detect the same pathogen in seafoods and other environmental samples. In this study, we detected and enumerated tdh gene-positive V. parahaemolyticus in Japanese seafoods with a tdh-specific PCR method, a chromogenic agar medium, and a most-probable-number method. The tdh gene was detected in 33 of 329 seafood samples (10.0%). The number of tdh-positive V. parahaemolyticus ranged from <3 to 93/10 g. The incidence of tdh-positive V. parahaemolyticus tended to be high in samples contaminated with relatively high levels of total V. parahaemolyticus. TDH-producing strains of V. parahaemolyticus were isolated from 11 of 33 tdh-positive samples (short-necked clam, hen clam, and rock oyster). TDH-producing strains of V. parahaemolyticus were also isolated from the sediments of rivers near the coast in Japan. Representative strains of the seafood and sediment isolates were examined for the O:K serovar and by the PCR method specific to the pandemic clone and arbitrarily primed PCR and pulsed-field gel electrophoresis techniques. The results indicated that most O3:K6 tdh-positive strains belonged to the pandemic O3:K6 clone and suggested that serovariation took place in the Japanese environment.
Figures


Similar articles
-
Epidemiological evidence of lesser role of thermostable direct hemolysin (TDH)-related hemolysin (TRH) than TDH on Vibrio parahaemolyticus pathogenicity.Foodborne Pathog Dis. 2015 Feb;12(2):131-8. doi: 10.1089/fpd.2014.1810. Foodborne Pathog Dis. 2015. PMID: 25646967
-
Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan.J Clin Microbiol. 1997 Dec;35(12):3150-5. doi: 10.1128/jcm.35.12.3150-3155.1997. J Clin Microbiol. 1997. PMID: 9399511 Free PMC article.
-
Major tdh(+)Vibrio parahaemolyticus serotype changes temporally in the Bay of Bengal estuary of Bangladesh.Infect Genet Evol. 2016 Jul;41:153-159. doi: 10.1016/j.meegid.2016.04.003. Epub 2016 Apr 7. Infect Genet Evol. 2016. PMID: 27063395
-
Structure, function and regulation of the thermostable direct hemolysin (TDH) in pandemic Vibrio parahaemolyticus.Microb Pathog. 2018 Oct;123:242-245. doi: 10.1016/j.micpath.2018.07.021. Epub 2018 Jul 19. Microb Pathog. 2018. PMID: 30031890 Review.
-
Pandemic Vibrio parahaemolyticus O3:K6 on the American continent.Front Cell Infect Microbiol. 2014 Jan 2;3:110. doi: 10.3389/fcimb.2013.00110. Front Cell Infect Microbiol. 2014. PMID: 24427744 Free PMC article. Review.
Cited by
-
Isolation and characterization of an antimicrobial Bacillus subtilis strain O-741 against Vibrio parahaemolyticus.PLoS One. 2024 Apr 4;19(4):e0299015. doi: 10.1371/journal.pone.0299015. eCollection 2024. PLoS One. 2024. PMID: 38573920 Free PMC article.
-
Characterization of Vibrio parahaemolyticus isolated from stool specimens of diarrhea patients in Nantong, Jiangsu, China during 2018-2020.PLoS One. 2022 Aug 26;17(8):e0273700. doi: 10.1371/journal.pone.0273700. eCollection 2022. PLoS One. 2022. PMID: 36018831 Free PMC article.
-
Changes in Volatile Composition of Cape Hake Fillets under Modified Atmosphere Packaging Systems during Cold Storage.Foods. 2022 Apr 29;11(9):1292. doi: 10.3390/foods11091292. Foods. 2022. PMID: 35564015 Free PMC article.
-
Recovery of Pasteurization-Resistant Vibrio parahaemolyticus from Seafoods Using a Modified, Two-Step Enrichment.Foods. 2022 Mar 7;11(5):764. doi: 10.3390/foods11050764. Foods. 2022. PMID: 35267397 Free PMC article.
-
Improved isolation and detection of toxigenic Vibrio parahaemolyticus from coastal water in Saudi Arabia using immunomagnetic enrichment.PeerJ. 2021 Oct 29;9:e12402. doi: 10.7717/peerj.12402. eCollection 2021. PeerJ. 2021. PMID: 34760388 Free PMC article.
References
-
- American Public Health Association. 1970. Recommended procedure for the examination of seawater and shellfish, 4th ed. American Public Health Association, Washington, D.C.
-
- Bag, P. K., S. Nandi, R. K. Bhadra, T. Ramamurthy, S. K. Bhattacharya, M. Nishibuchi, T. Hamabata, S. Yamasaki, Y. Takeda, and G. B. Nair. 1999. Clonal diversity among recently emerged strains of Vibrio parahaemolyticus O3:K6 associated with pandemic spread. J. Clin. Microbiol. 37:2354-2357. - PMC - PubMed
-
- Bhuiyan, N. A., M. Ansaruzzaman, M. Kamruzzaman, K. Alam, N. R. Chouwdhury, M. Nishibuchi, S. M. Faruque, D. A. Sack, Y. Takeda, and G. B. Nair. 2002. Prevalence of the pandemic genotype of Vibrio parahaemolyticus in Dhaka, Bangladesh, and significance of its distribution across different serotypes. J. Clin. Microbiol. 40:284-286. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources


