Pseudomonas fluorescens encodes the Crohn's disease-associated I2 sequence and T-cell superantigen
- PMID: 12438326
- PMCID: PMC133002
- DOI: 10.1128/IAI.70.12.6567-6575.2002
Pseudomonas fluorescens encodes the Crohn's disease-associated I2 sequence and T-cell superantigen
Abstract
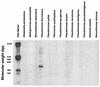
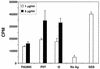
Commensal bacteria have emerged as an important disease factor in human Crohn's disease (CD) and murine inflammatory bowel disease (IBD) models. We recently isolated I2, a novel gene segment of microbial origin that is associated with human CD and that encodes a T-cell superantigen. To identify the I2 microorganism, BLAST analysis was used to identify a microbial homologue, PA2885, a novel open reading frame (ORF) in the Pseudomonas aeruginosa genome. PCR and Southern analysis identified Pseudomonas fluorescens as the originating species of I2, with homologues detectable in 3 of 13 other Pseudomonas species. Genomic cloning disclosed a locus containing the full-length I2 gene (pfiT) and three other orthologous genes, including a homologue of the pbrA/pvdS iron response gene. CD4(+) T-cell responses to recombinant proteins were potent for I2 and pfiT, but modest for PA2885. pfiT has several features of a virulence factor: association with an iron-response locus, restricted species distribution, and T-cell superantigen bioactivity. These findings suggest roles for pfiT and P. fluorescens in the pathogenesis of Crohn's disease.
Figures





References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Meyers, and D. J. Lippman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Auer, I. O., A. Roder, F. Wensinck, J. P. van de Merwe, and H. Schmidt. 1983. Selected bacterial antibodies in Crohn's disease and ulcerative colitis. Scand. J. Gastroenterol. 18:217-223. - PubMed
-
- Bhan, A. K., E. Mizoguchi, R. N. Smith, and A. Mizoguchi. 1999. Colitis in transgenic and knockout animals as models of human inflammatory bowel disease. Immunol. Rev. 169:195-207. - PubMed
-
- Blaser, M. J., R. A. Miller, J. Lacher, and J. W. Singleton. 1984. Patients with active Crohn's disease have elevated serum antibodies to antigens of seven enteric bacterial pathogens. Gastroenterology 87:888-894. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials


