Disruption of Brca2 increases the spontaneous mutation rate in vivo: synergism with ionizing radiation
- PMID: 11850397
- PMCID: PMC1084010
- DOI: 10.1093/embo-reports/kvf037
Disruption of Brca2 increases the spontaneous mutation rate in vivo: synergism with ionizing radiation
Abstract
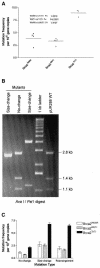
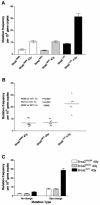
The breast cancer predisposition gene BRCA2 encodes a protein involved in the repair of DNA double-strand breaks, which arise spontaneously and following exposure to ionizing radiation (IR). To develop a mouse model that examines the effect of BRCA2 mutation and IR exposure on in vivo somatic mutation acquisition, we crossed mice with targeted disruption of Brca2 with a LacZ transgenic mutation reporter strain. Loss of both wild-type Brca2 alleles caused a 2.3-fold increase, equivalent to an extra 100 mutations per cell, in the in vivo acquisition of spontaneous somatic mutation by 2 weeks gestation. IR (4 Gy) had a disproportionate effect on animals homozygous for Brca2 disruption, inducing 3.4-fold more mutations compared with wild-type animals. These data provide the first evidence that loss of Brca2 increases in vivo somatic mutation acquisition and synergizes with IR exposure, with potential attendant implications for mammographic screening and therapeutic IR in mutation carriers.
Figures
Similar articles
-
Brca2 (XRCC11) deficiency results in enhanced mutagenesis.Mutagenesis. 2003 Nov;18(6):521-5. doi: 10.1093/mutage/geg032. Mutagenesis. 2003. PMID: 14614187
-
Spontaneous and irradiation-induced tumor susceptibility in BRCA2 germline mutant mice and cooperative effects with a p53 germline mutation.Toxicol Pathol. 2006;34(2):187-98. doi: 10.1080/01926230600611794. Toxicol Pathol. 2006. PMID: 16546942
-
Cell cycle and genetic background dependence of the effect of loss of BRCA2 on ionizing radiation sensitivity.Oncogene. 2003 May 15;22(19):2926-31. doi: 10.1038/sj.onc.1206522. Oncogene. 2003. PMID: 12771943
-
Diagnostic and therapeutic ionizing radiation and the risk of a first and second primary breast cancer, with special attention for BRCA1 and BRCA2 mutation carriers: a critical review of the literature.Cancer Treat Rev. 2015 Feb;41(2):187-96. doi: 10.1016/j.ctrv.2014.12.002. Epub 2014 Dec 8. Cancer Treat Rev. 2015. PMID: 25533736 Review.
-
Chromosomal mutagen sensitivity associated with mutations in BRCA genes.Cytogenet Genome Res. 2004;104(1-4):325-32. doi: 10.1159/000077511. Cytogenet Genome Res. 2004. PMID: 15162060 Review.
Cited by
-
Counting the cost of public and philanthropic R&D funding: the case of olaparib.J Pharm Policy Pract. 2022 Aug 16;15(1):47. doi: 10.1186/s40545-022-00445-9. J Pharm Policy Pract. 2022. PMID: 35974344 Free PMC article.
-
BRE/BRCC45 regulates CDC25A stability by recruiting USP7 in response to DNA damage.Nat Commun. 2018 Feb 7;9(1):537. doi: 10.1038/s41467-018-03020-6. Nat Commun. 2018. PMID: 29416040 Free PMC article.
-
BRCA1 and BRCA2 protect against oxidative DNA damage converted into double-strand breaks during DNA replication.DNA Repair (Amst). 2015 Jun;30:11-20. doi: 10.1016/j.dnarep.2015.03.002. Epub 2015 Mar 17. DNA Repair (Amst). 2015. PMID: 25836596 Free PMC article.
-
Tumour suppressor mechanisms in the control of chromosome stability: insights from BRCA2.Mol Cells. 2014 Feb;37(2):95-9. doi: 10.14348/molcells.2014.2346. Epub 2014 Feb 19. Mol Cells. 2014. PMID: 24598993 Free PMC article. Review.
-
BRCA2 and TP53 collaborate in tumorigenesis in zebrafish.PLoS One. 2014 Jan 29;9(1):e87177. doi: 10.1371/journal.pone.0087177. eCollection 2014. PLoS One. 2014. PMID: 24489863 Free PMC article.
References
-
- Bennett L.M. (1999) Breast cancer: genetic predisposition and exposure to radiation. Mol. Carcinog., 26, 143–149. - PubMed
-
- Boerrigter M.E., Dolle, M.E., Martus, H.J., Gossen, J.A. and Vijg, J. (1995) Plasmid-based transgenic mouse model for studying in vivo mutations. Nature, 377, 657–659. - PubMed
-
- Burke W. et al. (1997) Recommendations for follow-up care of individuals with an inherited predisposition to cancer. II. BRCA1 and BRCA2. Cancer Genetics Studies Consortium. JAMA, 277, 997–1003. - PubMed
-
- Connor F., Bertwistle, D., Mee, P.J., Ross, G.M., Swift, S., Grigorieva, E., Tybulewicz, V.L. and Ashworth, A. (1997) Tumorigenesis and a DNA repair defect in mice with a truncating Brca2 mutation. Nature Genet., 17, 423–430. - PubMed
-
- Dolle M.E., Martus, H.J., Gossen, J.A., Boerrigter, M.E. and Vijg, J. (1996) Evaluation of a plasmid-based transgenic mouse model for detecting in vivo mutations. Mutagenesis, 11, 111–118. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous


