Transduction by phiBB-1, a bacteriophage of Borrelia burgdorferi
- PMID: 11466280
- PMCID: PMC99531
- DOI: 10.1128/JB.183.16.4771-4778.2001
Transduction by phiBB-1, a bacteriophage of Borrelia burgdorferi
Abstract
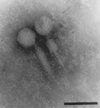
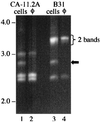
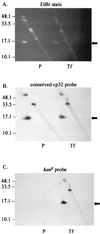
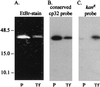
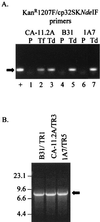
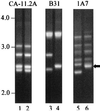
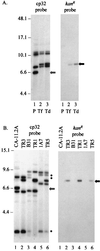
We previously described a bacteriophage of the Lyme disease agent Borrelia burgdorferi designated phiBB-1. This phage packages the host complement of the 32-kb circular plasmids (cp32s), a group of homologous molecules found throughout the genus Borrelia. To demonstrate the ability of phiBB-1 to package and transduce DNA, a kanamycin resistance cassette was inserted into a cloned fragment of phage DNA, and the resulting construct was transformed into B. burgdorferi CA-11.2A cells. The kan cassette recombined into a resident cp32 and was stably maintained. The cp32 containing the kan cassette was packaged by phiBB-1 released from this B. burgdorferi strain. phiBB-1 has been used to transduce this antibiotic resistance marker into naive CA-11.2A cells, as well as two other strains of B. burgdorferi. This is the first direct evidence of a mechanism for lateral gene transfer in B. burgdorferi.
Figures

Similar articles
-
Molecular evidence for a new bacteriophage of Borrelia burgdorferi.J Bacteriol. 1999 Dec;181(23):7308-13. doi: 10.1128/JB.181.23.7308-7313.1999. J Bacteriol. 1999. PMID: 10572135 Free PMC article.
-
Bacteriophages of spirochetes.J Mol Microbiol Biotechnol. 2000 Oct;2(4):365-73. J Mol Microbiol Biotechnol. 2000. PMID: 11075907 Review.
-
Phage-mediated horizontal gene transfer of both prophage and heterologous DNA by ϕBB-1, a bacteriophage of Borrelia burgdorferi.Pathog Dis. 2016 Dec;74(9):ftw107. doi: 10.1093/femspd/ftw107. Epub 2016 Nov 2. Pathog Dis. 2016. PMID: 27811049
-
Phage-Mediated Genetic Manipulation of the Lyme Disease Spirochete Borrelia burgdorferi.J Vis Exp. 2022 Sep 28;(187). doi: 10.3791/64408. J Vis Exp. 2022. PMID: 36279530
-
Repetition, conservation, and variation: the multiple cp32 plasmids of Borrelia species.J Mol Microbiol Biotechnol. 2000 Oct;2(4):411-22. J Mol Microbiol Biotechnol. 2000. PMID: 11075913 Review.
Cited by
-
Hitchhiker's Guide to Borrelia burgdorferi.J Bacteriol. 2024 Sep 19;206(9):e0011624. doi: 10.1128/jb.00116-24. Epub 2024 Aug 14. J Bacteriol. 2024. PMID: 39140751 Free PMC article. Review.
-
Characterization and genomic analysis of the Lyme disease spirochete bacteriophage ϕBB-1.PLoS Pathog. 2024 Apr 1;20(4):e1012122. doi: 10.1371/journal.ppat.1012122. eCollection 2024 Apr. PLoS Pathog. 2024. PMID: 38558079 Free PMC article.
-
Characterization and genomic analysis of the Lyme disease spirochete bacteriophage ϕBB-1.bioRxiv [Preprint]. 2024 Jan 13:2024.01.08.574763. doi: 10.1101/2024.01.08.574763. bioRxiv. 2024. Update in: PLoS Pathog. 2024 Apr 1;20(4):e1012122. doi: 10.1371/journal.ppat.1012122 PMID: 38260690 Free PMC article. Updated. Preprint.
-
Erp and Rev Adhesins of the Lyme Disease Spirochete's Ubiquitous cp32 Prophages Assist the Bacterium during Vertebrate Infection.Infect Immun. 2023 Mar 15;91(3):e0025022. doi: 10.1128/iai.00250-22. Epub 2023 Feb 28. Infect Immun. 2023. PMID: 36853019 Free PMC article. Review.
-
Coupled induction of prophage and virulence factors during tick transmission of the Lyme disease spirochete.Nat Commun. 2023 Jan 13;14(1):198. doi: 10.1038/s41467-023-35897-3. Nat Commun. 2023. PMID: 36639656 Free PMC article.
References
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1999.
-
- Burgdorfer W, Barbour A G, Hayes S F, Benach J L, Grunwaldt E, Davis J P. Lyme disease–a tick-borne spirochetosis? Science. 1982;216:1317–1319. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources


