Rare germinal unequal crossing-over leading to recombinant gene formation and gene duplication in Arabidopsis thaliana
- PMID: 10468603
- PMCID: PMC17883
- DOI: 10.1073/pnas.96.18.10302
Rare germinal unequal crossing-over leading to recombinant gene formation and gene duplication in Arabidopsis thaliana
Abstract
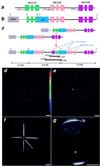
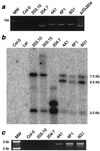
Small, multigene families organized in a tandem array can facilitate the rapid evolution of the gene cluster by a process of meiotic unequal crossing-over. To study this process in a multicellular organism, we created a synthetic RBCSB gene cluster in Arabidopsis thaliana and used this to measure directly the frequency of meiotic, intergenic unequal crossing-over between sister chromatids. The synthetic RBCSB gene cluster was composed of a silent DeltaRBCS1B::LUC chimeric gene fusion, lacking all 5' transcription and translation signals, followed by RBCS2B and RBC3B genomic DNA. Expression of luciferase activity (luc(+)) required a homologous recombination event between the DeltaRBCS1B::LUC and the RBCS3B genes, yielding a novel recombinant RBCS3B/ 1B::LUC chimeric gene whose expression was driven by RBCS3B 5' transcription and translation signals. Using sensitive, single-photon-imaging equipment, three luc(+) seedlings were identified in more than 1 million F2 seedlings derived from self-fertilized F1 plants hemizygous for the synthetic RBCSB gene cluster. The F2 luc(+) seedlings were isolated, and molecular and genetic analysis indicated that the luc(+) trait was caused by the formation of a recombinant chimeric RBCS3B/1B::LUC gene. A predicted duplication of the RBCS2B gene also was present. The recombination resolution break points mapped adjacent to a region of intron I at which a disjunction in sequence similarity between RBCS1B and RBCS3B occurs; this provided evidence supporting models of gene cluster evolution by exon-shuffling processes. In contrast to most measures of meiotic unequal crossing-over that require the deletion of a gene in a gene cluster, these results directly measured the frequency of meiotic unequal crossing-over (approximately 3 x 10(-6)), leading to the expansion of the gene cluster and the formation of a novel recombinant gene.
Figures

Similar articles
-
Meiotic recombination between paralogous RBCSB genes on sister chromatids of Arabidopsis thaliana.Genetics. 2004 Feb;166(2):947-57. doi: 10.1534/genetics.166.2.947. Genetics. 2004. PMID: 15020479 Free PMC article.
-
Frequency and character of alternative somatic recombination fates of paralogous genes during T-DNA integration.Mol Genet Genomics. 2005 Sep;274(2):91-102. doi: 10.1007/s00438-005-0001-z. Epub 2005 Oct 11. Mol Genet Genomics. 2005. PMID: 15983820
-
A gene duplication/loss event in the ribulose-1,5-bisphosphate-carboxylase/oxygenase (rubisco) small subunit gene family among accessions of Arabidopsis thaliana.Mol Biol Evol. 2011 Jun;28(6):1861-76. doi: 10.1093/molbev/msr008. Epub 2011 Jan 10. Mol Biol Evol. 2011. PMID: 21220760
-
Mice and the role of unequal recombination in gene-family evolution.Am J Hum Genet. 1999 Jan;64(1):40-5. doi: 10.1086/302220. Am J Hum Genet. 1999. PMID: 9915941 Free PMC article. Review. No abstract available.
-
Unequal crossing over then and now.Genetics. 1988 Sep;120(1):1-6. doi: 10.1093/genetics/120.1.1. Genetics. 1988. PMID: 3065137 Free PMC article. Review. No abstract available.
Cited by
-
Blessing or curse: how the epigenetic resolution of host-transposable element conflicts shapes their evolutionary dynamics.Proc Biol Sci. 2024 Apr 10;291(2020):20232775. doi: 10.1098/rspb.2023.2775. Epub 2024 Apr 10. Proc Biol Sci. 2024. PMID: 38593848 Free PMC article.
-
Natural and artificial sources of genetic variation used in crop breeding: A baseline comparator for genome editing.Front Genome Ed. 2022 Aug 22;4:937853. doi: 10.3389/fgeed.2022.937853. eCollection 2022. Front Genome Ed. 2022. PMID: 36072906 Free PMC article. Review.
-
A 69 kbp Deletion at the Berry Color Locus Is Responsible for Berry Color Recovery in Vitis vinifera L. Cultivar 'Riesling Rot'.Int J Mol Sci. 2022 Mar 28;23(7):3708. doi: 10.3390/ijms23073708. Int J Mol Sci. 2022. PMID: 35409066 Free PMC article.
-
Novel Structural Variation and Evolutionary Characteristics of Chloroplast tRNA in Gossypium Plants.Genes (Basel). 2021 May 27;12(6):822. doi: 10.3390/genes12060822. Genes (Basel). 2021. PMID: 34071968 Free PMC article.
-
Poison ivy hairy root cultures enable a stable transformation system suitable for detailed investigation of urushiol metabolism.Plant Direct. 2020 Aug 7;4(8):e00243. doi: 10.1002/pld3.243. eCollection 2020 Aug. Plant Direct. 2020. PMID: 32783021 Free PMC article.
References
-
- Ruddle F H, Bartels J L, Bentley K L, Kappen C, Murtha M T, Pendleton J W. Annu Rev Genet. 1994;28:423–442. - PubMed
-
- Collins F S, Weissman S M. Prog Nucleic Acid Res Mol Biol. 1984;31:315–462. - PubMed
-
- Hughes A L, Yeager M. BioEssays. 1997;19:777–786. - PubMed
-
- Matsuda F, Honjo T. In: Advances in Immunology. Dixon F J, editor. Vol. 62. San Diego: Academic; 1996. pp. 1–29. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous


