Abstract
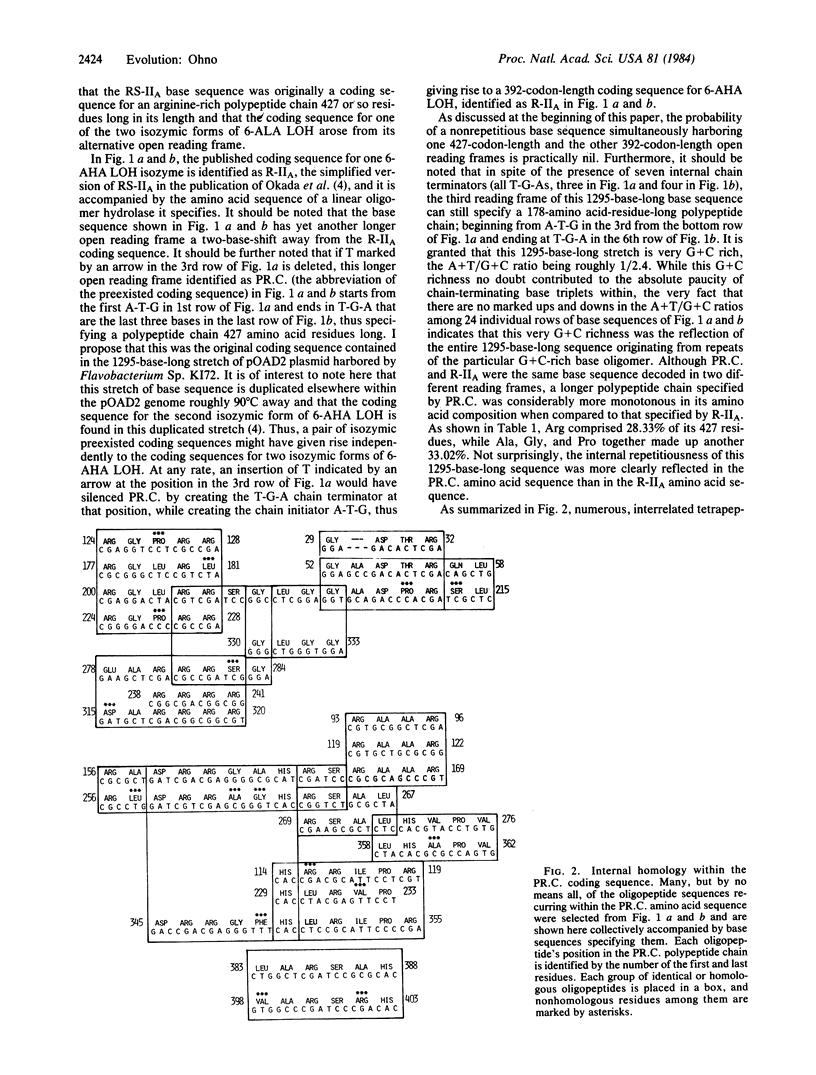
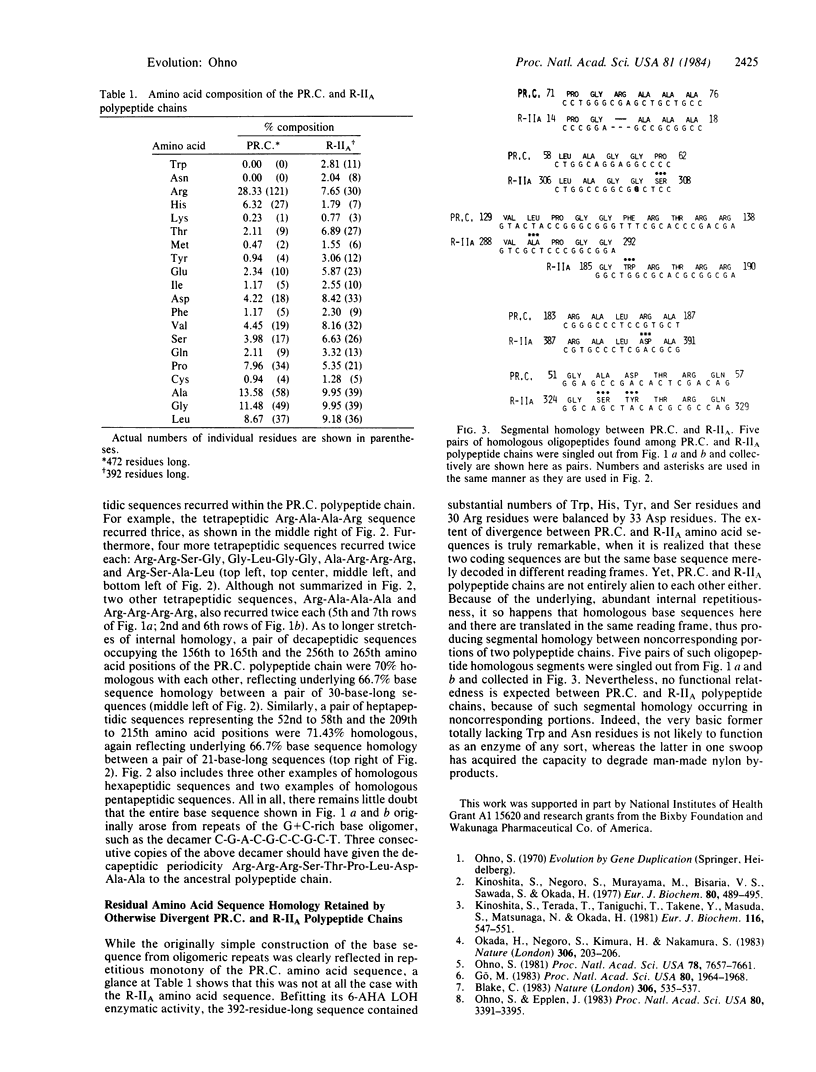
The mechanism of gene duplication as the means to acquire new genes with previously nonexistent functions is inherently self limiting in that the function possessed by a new protein, in reality, is but a mere variation of the preexisted theme. As the source of a truly unique protein, I suggest an unused open reading frame of the existing coding sequence. Only those coding sequences that started from oligomeric repeats are likely to retain alternative long open reading frames. Analysis of the published base sequence residing in the pOAD2 plasmid of Flavobacterium Sp. K172 indicated that the 392-amino acid-residue-long bacterial enzyme 6-aminohexanoic acid linear oligomer hydrolase involved in degradation of nylon oligomers is specified by an alternative open reading frame of the preexisted coding sequence that originally specified a 472-residue-long arginine-rich protein.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blake C. Exons--present from the beginning? Nature. 1983 Dec 8;306(5943):535–537. doi: 10.1038/306535a0. [DOI] [PubMed] [Google Scholar]
- Go M. Modular structural units, exons, and function in chicken lysozyme. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1964–1968. doi: 10.1073/pnas.80.7.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita S., Negoro S., Muramatsu M., Bisaria V. S., Sawada S., Okada H. 6-Aminohexanoic acid cyclic dimer hydrolase. A new cyclic amide hydrolase produced by Achromobacter guttatus KI74. Eur J Biochem. 1977 Nov 1;80(2):489–495. doi: 10.1111/j.1432-1033.1977.tb11904.x. [DOI] [PubMed] [Google Scholar]
- Kinoshita S., Terada T., Taniguchi T., Takene Y., Masuda S., Matsunaga N., Okada H. Purification and characterization of 6-aminohexanoic-acid-oligomer hydrolase of Flavobacterium sp. Ki72. Eur J Biochem. 1981 Jun 1;116(3):547–551. doi: 10.1111/j.1432-1033.1981.tb05371.x. [DOI] [PubMed] [Google Scholar]
- Ohno S., Epplen J. T. The primitive code and repeats of base oligomers as the primordial protein-encoding sequence. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3391–3395. doi: 10.1073/pnas.80.11.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Original domain for the serum albumin family arose from repeated sequences. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7657–7661. doi: 10.1073/pnas.78.12.7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H., Negoro S., Kimura H., Nakamura S. Evolutionary adaptation of plasmid-encoded enzymes for degrading nylon oligomers. Nature. 1983 Nov 10;306(5939):203–206. doi: 10.1038/306203a0. [DOI] [PubMed] [Google Scholar]



