Abstract
The regulation of telomere length in mammals is crucial for chromosome end-capping and thus for maintaining genome stability and cellular lifespan. This process requires coordination between telomeric protein complexes and the ribonucleoprotein telomerase, which extends the telomeric DNA. Telomeric proteins modulate telomere architecture, recruit telomerase to accessible telomeres and orchestrate the conversion of the newly synthesized telomeric single-stranded DNA tail into double-stranded DNA. Dysfunctional telomere maintenance leads to telomere shortening, which causes human diseases including bone marrow failure, premature ageing and cancer. Recent studies provide new insights into telomerase-related interactions (the ‘telomere replisome’) and reveal new challenges for future telomere structural biology endeavours owing to the dynamic nature of telomere architecture and the great number of structures that telomeres form. In this Review, we discuss recently determined structures of the shelterin and CTC1–STN1–TEN1 (CST) complexes, how they may participate in the regulation of telomere replication and chromosome end-capping, and how disease-causing mutations in their encoding genes may affect specific functions. Major outstanding questions in the field include how all of the telomere components assemble relative to each other and how the switching between different telomere structures is achieved.
Unlike circular chromosomes, linear chromosomes require a special mechanism to preserve their genetic material during DNA replication and also to prevent false recognition of the chromosome ends as DNA breakage sites. The telomere, which is composed of repetitive DNA sequences and specialized proteins, serves this purpose by providing a protective cap to chromosome ends. In humans, the telomeric DNA sequence is (TTAGGG)n (REF.1) and is evolutionarily conserved across vertebrates2. In addition to its repetitive DNA property, the telomere has an uncommon DNA architecture comprising two structures: a double-stranded DNA (dsDNA) region several kilobases long, ending with a single-stranded 3′ tail known as the G-overhang3 (FIG. 1). The length of telomeres is highly heterogeneous4, even within a single cell, and serves as a ‘molecular clock’ of the proliferative lifespan of primary cells as, in the absence of extension, telomere length is progressively shortened in every cell division5–7. Critically short telomeres can trigger cell entry to cellular senescence8, also known as the Hayflick limit of proliferative lifespan. The initial length of telomeres in somatic cells is derived from germline cells, in which telomere length is actively maintained. Hence, telomere length maintenance has an important role in cell biology, and its deregulation can lead to premature ageing and cancer9–12.
Fig. 1 |. Telomere DNA structures at chromosome ends.
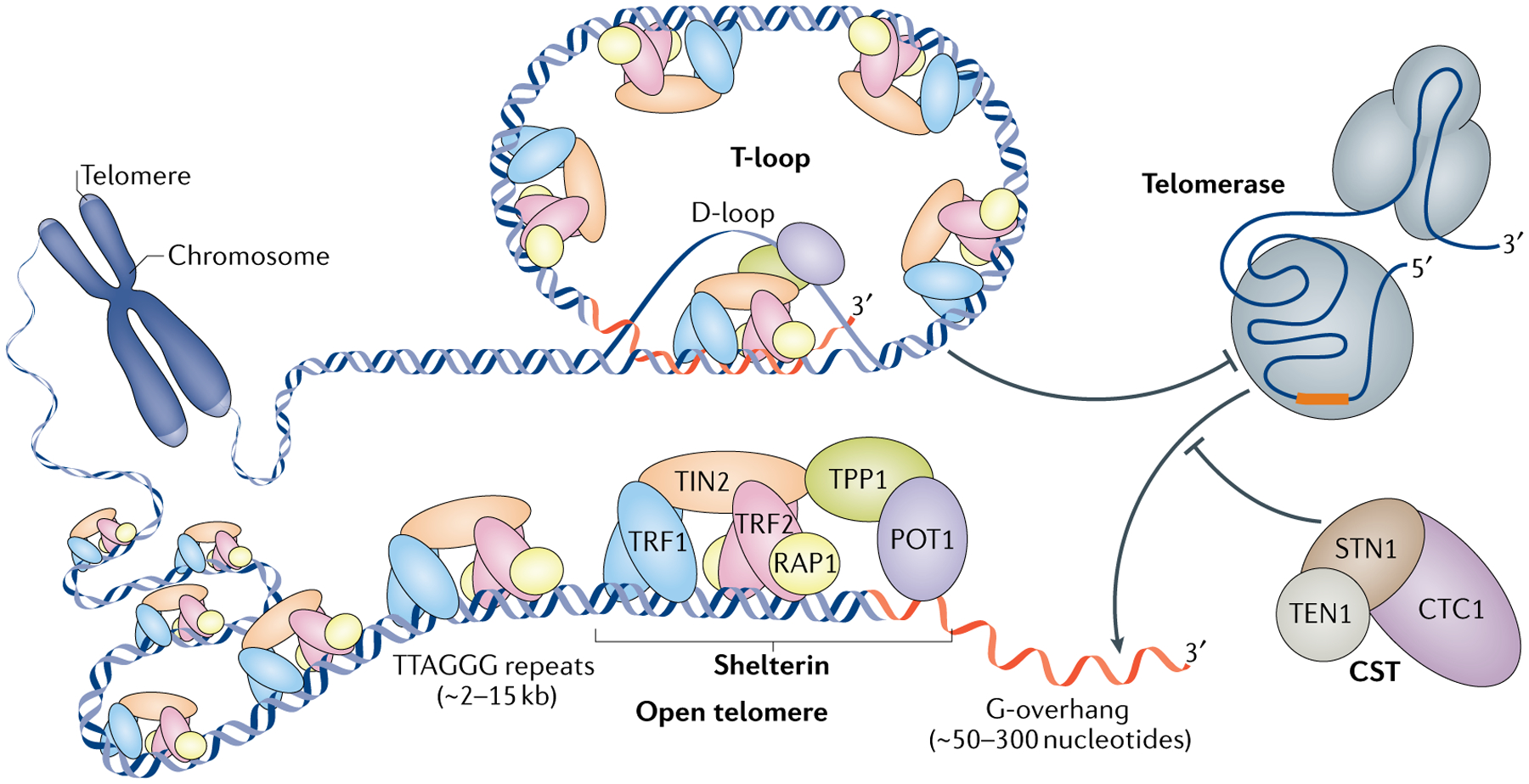
Telomeres are DNA–protein structures at the ends of linear chromosomes. Depicted are the essential telomeric protein complexes shelterin and CTC1–STN1–TEN1 (CST), as well the telomerase RNA–protein complex. The telomeric DNA consists of both double-stranded DNA and single-stranded DNA, comprising in vertebrates the repeat sequence TTAGGG. This duality allows the single-stranded 3′ tail (red) to invade the double-stranded DNA region to form a displacement loop (D-loop) and a telomere loop (T-loop). T-loop formation is regulated by shelterin complexes and can restrict telomerase access to the 3′ tail. Once the telomere is opened up — presumably during the S phase of the cell cycle — telomerase can bind to the 3′ tail through its RNA template (orange) and add telomeric repeats. CST then inhibits telomerase activity, thereby preventing excessive telomere extension. POT1, protection of telomeres protein 1; TIN2, TERF1-interacting nuclear factor 2; TRF1, telomeric repeat-binding factor 1.
G-overhang.
A telomeric single-stranded DNA 3′ tail consisting of TTAGGG repeats, which is the substrate of telomerase.
Shelterin (also called telosome) is a protein complex comprising up to six different proteins; the composition of the complex may vary depending on the chromosomal context. Shelterin binds both the single-stranded DNA (ssDNA) and dsDNA regions of telomeres13–16, and is crucial for both telomere protection and telomerase regulation. Because of its unique DNA-binding nature, the shelterin complex can organize telomeres into various protein–DNA structures, such as the end-capped telomere17,18 or the telomere loop (T-loop)19–22, which effectively hides (and thus protects) the telomeric tail through strand invasion into the double-stranded region (FIG. 1). Owing to the repetitive nature of telomeric DNA, the association of multiple shelterin and other telomeric protein complexes with the telomere has implications for how cells sense and respond to telomere length changes23,24. It seems likely that the telomere higher-order architecture must be unravelled for the telomeric tail to be rendered accessible to telomerase.
Telomerase uses its intrinsic RNA template to synthesize telomeric DNA repeats25,26 and adds ~60 nucleotides per telomere per cell cycle in a processive manner27 (for reviews of telomerase, see REFS28–30). In most cases in humans, there is a major difference in the expression of telomerase between somatic cells and germline cells: telomerase is more abundant in germline cells31, which helps maintain telomere length, and is absent or less abundant in somatic cells11. Not surprisingly, immortalized cells and cancer cells often have high telomerase activity12. Telomerase recruitment to telomeres is mediated by the shelterin complex32–34. The timely termination of the telomere extension process is mediated by another telomeric protein complex, the heterotrimeric CTC1–STN1–TEN1 (CST) complex35–38. CST is also required for the recruitment of DNA polymerase α-primase (pol α-primase) to the newly synthesized telomeric tail for C-strand fill-in (conversion of part of the extended tail to dsDNA)39. Another macromolecular component of telomeres is the long non-coding RNA, telomeric repeat-containing RNA (TERRA)40. Collectively, telomere maintenance is a complex process regulated by large-scale chromatin architectural modulations and dynamic molecular interactions.
Recent developments in our understanding of the structural aspects of telomere maintenance and its dynamics prompt a timely evaluation of current models of telomere biology. In this Review, we discuss human telomere structure and replication in the telomerase-mediated pathway, including the latest structural information on two major telomeric protein complexes (shelterin and CST) and the dynamics of telomerase and telomeric proteins at telomeres. We propose an updated overview of various telomere regulation models, with the goal of reassessing long-standing questions and exploring new models for telomere maintenance.
Telomerase.
A ribonucleoprotein that uses its RNA as a template to synthesize TTAGGG repeats, thereby extending telomeres.
Processive.
The ability of an enzyme to perform multiple catalytic reactions without releasing its substrate.
Telomeric repeat-containing RNA.
(TERRA). A long non-coding RNA involved in regulating telomerase activity at telomeres.
Sheltering the chromosome end
The chief guardian of the chromosome ends is the six-member shelterin complex — comprising RAP1, telomeric repeat-binding factor 1 (TRF1), TRF2, TERF1-interacting nuclear factor 2 (TIN2), TPP1 and protection of telomeres protein 1 (POT1). Shelterin binds telomeric DNA in the context of nucleosomes, a constraint that is discussed below (see Other human telomere proteins). In addition to its end-protection function41, shelterin also provides a sensing mechanism of telomere length and controls the activity of telomerase at the telomere17,42–47. TRF1 and TRF2 are homodimers that bind telomeric dsDNA48–51, whereas the POT1–TPP1 heterodimer binds and caps the telomeric 3′ tail17,18,52. TIN2 connects TRF1 and TRF2 dimers with POT1–TPP1 to form the shelterin complex14,15,53 (FIG. 2a). RAP1 is an accessory subunit of TRF2 that contributes to telomere protection54,55, specifically at critically short telomeres56.
Fig. 2 |. Molecular architecture of the human shelterin complex.
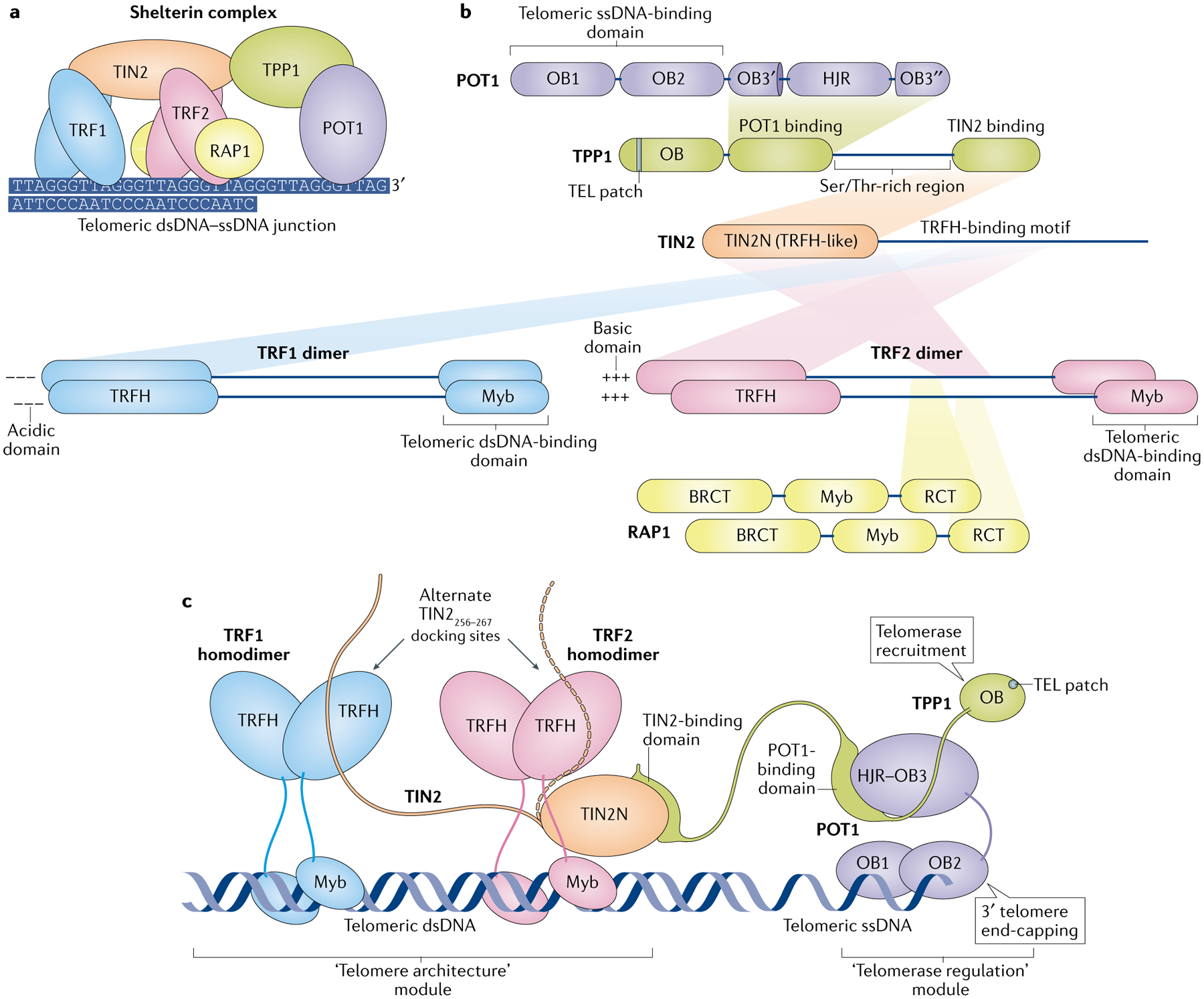
a | The human shelterin complex consists of six proteins: telomeric repeat-binding factor 1 (TRF1), TRF2, RAP1, TERF1-interacting nuclear factor 2 (TIN2), TPP1 and protection of telomeres protein 1 (POT1). Shelterin engages both double-stranded DNA (dsDNA) and single-stranded DNA (ssDNA) regions of a telomere. TRF1 and TRF2 are protein homodimers that bind telomeric dsDNA, whereas POT1 binds telomeric ssDNA. b | Map of interactions of shelterin subunits, with binding and structured domains illustrated as boxes. Each polypeptide is displayed from the amino terminus (left) to the carboxy terminus (right) and their stoichiometry is indicated by the number of copies. c | Possible organization of an assembled shelterin complex (excluding RAP1). Unstructured regions are illustrated as coloured lines, depicting the large flexible regions of TRF1 and TRF2 (between their TRF homology (TRFH) domains and Myb domains) and of TPP1 (the C-terminal region after its oligonucleotide/oligosaccharide-binding (OB) domain). The fully assembled shelterin complex comprises two structurally separated modules, which we term the telomere architecture module (TRF1–TIN2–TRF2–RAP1) and the telomerase regulation module (TPP1–POT1). A single TIN2 (TIN2256–276) can interact with the TRFH domain of either TRF1 or TRF2 (dashed arm), whereas the TIN2 N-terminal domain (TIN2N) interacts solely with TRF2. BRCT, BRCA1 carboxy terminal; HJR, Holliday junction resolvase-like; RCT, RAP1 carboxy terminal; TEL patch, TPP1 glutamate (E) and leucine (L)-rich patch.
Organization and DNA-binding properties
The majority of shelterin subunits interact minimally with other subunits of the complex, with each subunit having a domain that interacts with adjacent subunit(s) to form a branched structure (FIG. 2a). The telomeric ssDNA-binding domain of shelterin lies mainly in the amino-terminal OB1 and OB2 domains of POT1 (REF.18) (FIG. 2b), and the addition of TPP1 increases the affinity of POT1 for DNA tenfold57,58. (An OB domain or OB-fold domain is a small oligonucleotide-binding or oligosaccharide-binding structural motif found in many proteins.) Both POT1 by itself and POT1–TPP1 have a preference for capping the telomeric 3′ end, but they also bind to internal sites of telomeric ssDNA59. POT1 interacts with amino acids 266–320 of TPP1 (TPP1266–320) through two structured domains (across POT1 OB3 and Holliday junction resolvase-like (HJR) domains)60,61, with no known interactions with other shelterin subunits. The discovery of an HJR domain in POT1 (REFS60,61), which is structurally adjacent to its telomeric ssDNA-binding domain (FIG. 2c), is intriguing given the possibility that a T-loop can progress to become a telomeric Holliday junction62. The TPP1 N-terminal OB domain contains the TPP1 glutamate (E) and leucine (L)-rich patch (TEL patch), which recruits telo merase to telomeres32–34, whereas its carboxy terminus (TPP1510–544)15,46,53 interacts with the TIN2 N terminus63,64 (TIN2N). TIN2, which is effectively the hub of shelterin assembly, links TPP1 with TRF1 and TRF2 homodimers. The TIN2N domain, which structurally resembles the TRF homology (TRFH) domains of TRF1 and TRF2, interacts with both TPP1 and TRF2350–366 (REF.63). In addition, another region of TIN2 (TIN2256–276) interacts with the TRF2 TRFH domain65. Interestingly, whereas the TRF1 and TRF2 TRFH domains are structurally similar66, they do not share a similar level of interaction with TIN2 (REFS43,65): the TRFH domain of TRF1 binds TIN2256–276 20-fold tighter than the TRFH domain of TRF2 (REF.65).
Holliday junction.
A branched DNA structure comprising four double-stranded arms.
TEL patch.
(TPP1 glutamate (E) and leucine (L)-rich patch).
A small group of amino acids on the surface of the shelterin protein TPP1. The TEL patch directly recruits telomerase to telomeres and then stimulates its activity.
Both TRF1 and TRF2 homodimers have a Myb DNA-binding domain at their C terminus for sequence-specific binding of telomeric dsDNA (YTAGGGTTR; Y = C or T, R = A or G)48,49,51,67–70. The TRFH domain is connected to the Myb domain by a long polypeptide chain (~100 amino acids in TRF1 and ~200 amino acids in TRF2), which has no apparent structural features; this flexibility allows each Myb domain of a TRF1 homodimer or of a TRF2 homodimer to bind telomeric DNA with various distances and orientations with regards to the other Myb domain20,67,71 (FIG. 2c). This flexibility may be crucial for TRF1 and TRF2 binding chromatin comprising nucleosomes with short linker DNAs, as discussed below (see Other human telomere proteins).
TRF1 and TRF2 differ in their N termini. TRF11–67 is acidic (Asp–Glu-rich), whereas TRF21–45 is basic (Arg-rich). This difference translates to differences in how TRF1 and TRF2 interact with DNA or mediate the formation of special DNA architectures. The TRF1 N-terminal acidic domain has been implicated in the recruitment of the poly(ADP-ribose) polymerase tankyrase 1 (REF.72) and inhibition of telomeric DNA condensation73. The N-terminal basic domain of TRF2, which is better studied, is involved in the protection74 and regulation75,76 of T-loops, interaction with histones77 and inhibition of DNA double-strand break repair by non-homologous DNA end-joining in conjunction with RAP1 (REF.54). Given its positive charge, one might suggest that the basic domain participates in the interaction of TRF2 with canonical dsDNA such as telomeric DNA. However, the TRF2 N-terminal basic domain does not directly participate in TRF2 recruitment to telomeres but is instead involved in engaging non-canonical DNA structures such as T-loops78, Holliday junctions79 and G-quadruplexes (G4s)80–82. The proposed ability of RAP1 to tune the specificity of TRF2 for telomeric dsDNA by neutralizing the basic domain83 suggests that RAP1 has other roles in regulating the affinity of TRF2 to these non-canonical DNA structures.
Shelterin possesses the special ability to engage both telomeric ssDNA and dsDNA. Consequently, mammalian shelterin complexes consisting of POT1 with either TRF1 or TRF2 exhibit high affinity to telomeric DNA comprising both ssDNA and dsDNA regions53,84,85, and the minimal reconstituted human shelterin complex (shelterincore, consisting of TRF2–TIN2–TPP1–POT1) binds ssDNA–dsDNA junctions with nanomolar affinity53. Shelterincore binding to telomeric ssDNA–dsDNA junctions is not easily competed by telomeric ssDNA or dsDNA53. This provides the biochemical basis for how the shelterin complex could specifically target various telomeric DNA structures that present both ssDNA and dsDNA regions and provides insights into how shelterin recruits telomerase to the telomeric tail (for a detailed discussion, see Telomerase inhibition and stimulation).
G-quadruplexes.
(G4s). Tertiary structures in which groups of four guanines in single-stranded DNA form tetrads through hydrogen bonds.
Architecture of the shelterin complex
Although the atomic structures of several shelterin protein domains have been solved18,57,60,61,63,66,68,86,87, the overall architecture of mammalian shelterin as a complex remains unknown. Furthermore, two fundamental questions remain: which shelterin complexes and subcomplexes are functional in vivo, with what subunit stoichiometry; and with which telomeric DNA structures does each of them interact?
Functions and stoichiometry of shelterin complexes in vivo.
Shelterin has often been illustrated or described as a six-member protein complex; however, a minimum of four different shelterin complexes would be required to account for the uneven stoichiometry of chromatin-bound shelterin subunits in cells88. The most abundant of these complexes would contain two TRF2 subunits (that is, one dimer), two RAP1 subunits and one TIN2 subunit. The next most abundant is the complex of two TRF2 subunits, two RAP1 subunits, two TRF1 subunits and one TIN2 subunit. Finally, only about 10% of these dsDNA-binding complexes are associated with TPP1–POT1. Thus, the six-member complex comprises perhaps 5% of the shelterin complexes. The five-member shelterin complex missing only TRF1 has been reconstituted and purified, which allowed its molecular stoichiometry to be measured: two TRF2 subunits, two RAP1 subunits and one each of TIN2, TPP1 and POT1 subunits (REF.53). Moreover, the above discussion of different shelterin complexes underestimates the actual number, as it disregards the diversity provided by protein isoforms89–91 and post-translational modifications92,93. In any case, ‘shelterin’ is a population of related complexes, not a single molecular entity. These complexes may very well function differently in space (for example, position along the telomere) and time (for example, phase of the cell cycle).
Furthermore, the compositions of shelterin complexes are not static but dynamic. The dynamic aspect of human shelterin assembly has been shown with the ability of TRF1 to replace TRF2 in association with TIN2 and the conformational changes conferred by TPP1 incorporation, which lead to simultaneous accommodation of both TRF1 and TRF2 in a single shelterin complex13,94,95. The dynamic nature of shelterin is also consistent with the relatively weak (micromolar-range) affinity of interactions between shelterin proteins61,63,65,83, suggesting that the assembly of shelterin is likely dynamic and synergistic and that its composition is determined by contextual telomeric DNA structures and/or additional protein partners. The above considerations highlight the importance of directing future research to investigate shelterin functions and dynamics in the context of complexes rather than individual proteins.
Perhaps underappreciated features of shelterin are the several long flexible regions connecting the structured domains (FIG. 2b,c). In TPP1, between the TIN2-binding site and the POT1-binding site, there is a Ser/Thr-rich region that contains several highly conserved Ser/Thr phosphorylation motifs96. The function of the Ser/Thr-rich region of TPP1 is still unclear, but given that it bridges TIN2 and POT1, its potential phosphorylation states might yet have implications for shelterin architecture or telomere organization. Its length of ~150 amino acids with no apparent secondary structures (from in silico prediction) suggests that the Ser/Thr-rich region behaves like a long flexible peptide chain that would effectively segregate TPP1 and POT1 from the rest of the shelterin proteins (FIG. 2c). We refer to the TRF1–TRF2–RAP1–TIN2 and TPP1–POT1 subcomplexes as the ‘telomere architecture’ module and the ‘telomerase regulation’ module, respectively, based on their functions. As already mentioned, both TRF1 and TRF2 also have a long unstructured region that connects their TRFH and Myb domains, which confers further conformational flexibility and adaptability to how shelterin assembles itself on telomeric DNA. It is thus highly possible that shelterin leverages this flexibility to exhibit degenerate affinity to various telomeric DNA structures such as the T-loop or Holliday junctions, or the telomeric ssDNA–dsDNA junction at the very end of the chromosome.
With which telomeric DNA structures do various shelterin complexes interact.
Early biochemical studies focused on mammalian telomere proteins binding to linear telomeric DNA, either dsDNA in the case of TRF1 and TRF2 or ssDNA in the case of POT1–TPP1. However, considering the flexibility of the complexes and their functional interactions with diverse telomeric DNA structures, it is important to investigate and compare shelterin complex affinities with multiple DNA structures. The simplest of these is a linear dsDNA with a 3′ ssDNA tail53,84,85, as had been studied earlier with ciliate telomere junctions97. A more complex structure is the displacement loop (D-loop) at the base of the T-loop (FIG. 1), in which the invading telomeric single strand (red) displaces an equivalent portion of the G-rich strand, thereby presenting both telomeric ssDNA and dsDNA in close proximity. It is thus often proposed and illustrated that an assembled shelterin complex (containing TRFs and POT1 proteins) would help stabilize or mediate D-loop formation98. It has been shown in vivo that TRF2 is the dominant factor driving T-loop (or D-loop) formation22,99, and, consistent with that observation, TRF2 is sufficient to drive T-loop formation in vitro20. However, it will be important to understand how shelterin complexes compare with TRF2 in regulating T-loop formation. For example, there remains a possible cooperative effect of POT1 (in the context of a shelterin complex) in stabilizing T-loops. Also, TPP1 association with POT1 can elevate the ssDNA binding affinity of POT1 (REF.57) and RAP1 association with TRF2 affects TRF2 binding to dsDNA83, both studies suggesting that higher-order assembly of shelterin complexes is accompanied by allosteric changes that modulate their affinities to telomeric DNA. Hence, it will be important for future research to investigate how the various components of shelterin interact with telomeric D-loops (T-loops)100, Holliday junctions76 and linear telomeres of various geometries.
High-resolution structures of assembled shelterin complexes, either free or bound to key telomeric DNA substrates, remain elusive. Cryo-electron microscopy (cryo-EM) has revolutionized structural biology101,102 and seems likely to provide the breakthrough needed. The structural biology of telomerase has greatly benefited from the electron microscopy reconstruction approach103,104 whereas shelterin cryo-EM has not seen much progress. This could be owing to the intrinsic conformational flexibility and dynamic assembly of shelterin complexes. However, given that many structured domains of shelterin have been solved by X-ray crystallography18,57,60,61,63,66,68,86,87, a medium-resolution cryo-EM structure of a mammalian shelterin complex should allow docking of these solved structures into the overall envelope; this would yield many insights into shelterin architecture and organization, as shown by the human telomerase cryo-EM structure104. Future development of cryo-EM software that is able to accommodate samples with high conformational heterogeneity will be an important step towards progressing shelterin structural biology.
Telomerase inhibition and stimulation
Shelterin has multiple roles in regulating telomerase activity at telomeres. On the one hand, shelterin can mediate T-loop formation20,22 or end-cap the telomeric 3′ tail17,47,52,59 to restrict telomerase access. On the other hand, it can recruit telomerase to the telomere32–34 and drive telomerase processivity53,57. The recruitment of telomerase to the telomere by shelterin is mediated directly by the TEL patch on the TPP1 OB domain32–34. The recruitment process occurs in the S phase of the cell cycle, is highly dynamic and involves multiple kinetically distinct stages. Single-molecule live-cell imaging of human telomerase reverse transcriptase (hTERT; the catalytic protein component of telomerase) in cells revealed that telomerase samples telomeres multiple times using a 3D diffusive search mechanism, before stably engaging the telomeric tail through base pairing with the template region of the telomerase RNA (hTR) component105,106; both the probing interactions and stable binding require interaction with the TEL patch. Similar experiments with hTR corroborated the hTERT observations and also revealed early stages of hTR biogenesis and telomerase assembly107.
Although the direct role of TPP1 in telomerase recruitment is starting to be understood, its own recruitment to telomeres and assembly with the other components of shelterin are less clear. In vitro studies show mouse shelterin complexes sampling the DNA in a 3D search before stably engaging telomeric sequences85. Another study has shown that human TRF1 and TRF2 exhibit slower diffusive behaviour on relatively short regions of telomeric DNA108. Clearly, the mobility of shelterin on telomeric DNA and on telomeric chromatin, including sliding and hopping, are important questions for the future. For example, given the repetitive sequence of the telomere, how does TPP1 help telomerase find the telomeric tail instead of ‘distracting’ it to internal sites? The DNA sequence might be the same but the structure at the end of the telomere is unique, where both telomeric ssDNA and dsDNA exist (FIG. 1). Hence, the answer might lie in the necessity of an assembled shelterin complex consisting of TRF1 and/or TRF2, and POT1, which would direct the complex to search for the telomeric ssDNA–dsDNA junction based on the virtue of highest binding affinity53 and eventually directing telomerase to the telomeric end.
Interestingly, despite its high affinity to telomeric ssDNA, POT1 localization to telomeres in vivo depends on its association with TPP1 (REFS45,46,52), indicating that TPP1–POT1 likely exists as a heterodimer during its recruitment to telomeres. Nonetheless, the recruitment of TPP1 to the very end of the telomere may involve multiple stages. For example, as an initial step, TPP1–POT1 could transiently associate with TIN2–TRF1–TRF2 complexes that are bound along telomeric dsDNA regions, sliding or hopping along the telomeres until it finds the telomeric ssDNA–dsDNA junction; it would then form a particularly stable protein–DNA complex, in which TPP1–POT1 is bound to the ssDNA and the TRFs are bound to the dsDNA. Whether TPP1–POT1 located at the ssDNA–dsDNA junction is close enough to the DNA 3′ end to load telomerase remains to be determined.
A particular conundrum is how shelterin switches between its inhibitory and stimulatory roles in regulating telomerase activity at telomeres. Insights might come from the dynamic multiple activities of POT1: it is essential for telomeric ssDNA protection by preventing aberrant accumulation of replication protein A (RPA)109,110; it participates in cell-cycle coordinated RPA removal through TERRA and heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1)111; and it associates with TPP1 to stimulate telomerase processivity57. POT1 function depends on its positioning on telomeric ssDNA. For example, POT1 binding to internal telomeric ssDNA allows telomerase to bind to the DNA 3′ end, whereas POT1 binding to the 3′ end of telomeric ssDNA excludes telomerase59. The 3′ end binding by POT1–TPP1 gives telomere protection, whereas the internal binding allows telomerase recruitment and stimulation.
However, the role of POT1 extends beyond just positioning, as it is also able to disrupt G4 formation by telomeric ssDNA112,113, which otherwise can exclude telomerase binding113,114. The DNA-binding behaviour of POT1 is further changed upon association with TPP1, causing POT1 (together with TPP1) to slide back and forth along the ssDNA to iron out G4 structures115. But is G4 always an inhibitory structure to telomerase? Apparently not. Recent studies have shown that a G4 is formed in nascent telomeric ssDNA synthesized by telomerase and can aid telomerase processivity116,117. It was further proposed that TPP1–POT1 interaction with this G4 can facilitate the fidelity of telomerase translocation and nascent DNA protection116, which would contribute to TPP1–POT1 stimulation of telomerase processivity118. The intermediate-resolution model of human telomerase104 already suggests the existence of a cavity to house this G4 (REF.116). More recently, single-molecule fluorescence studies have shown that the telomerase RNA template can directly interact with parallel-stranded G4 structures and facilitate G4 disruption upon telomerase translocation119. A high-resolution structure of telomerase in conjunction with shelterin complexes would provide more insights into these complex processes.
Replication protein A.
(RPA). A eukaryotic single-stranded DNA-binding protein complex involved in DNA replication and repair.
Telomeric C-strand.
The telomeric DNA strand consisting of CCCTAA repeats, which pair with the telomerase-synthesized TTAGGG repeats.
Switching to synthesis of the C-strand
Mammalian CST heterotrimeric complex (FIG. 3a,b) was identified as a telomeric component based on homology to the yeast CST complex (Cdc13–Stn1–Ten1)36; both complexes possess similarity to the RPA heterotrimer in their OB domain predictions and ssDNA-binding properties37,38. Like yeast CST, mammalian CST participates in the replication and maintenance of telomeres. The depletion of CTC1 or STN1 results in telomeric tail (G-overhang) lengthening38,120,121. This is owing to failure of the telomeric C-strand fill-in process, which depends on the recruitment and action of pol α-primase to convert a portion of the newly synthesized telomeric tail to dsDNA form. Because of its ssDNA-binding property37, CST can compete with TPP1–POT1 for binding the telomeric 3′ tail and also sequester telomerase from telomeric tail engagement35. This ssDNA-binding property allows CST to terminate the extension of the telomere by telomerase during the late S/G2 cell cycle phase35 and then facilitate C-strand fill-in. Emerging roles of CST have expanded its territory beyond telomeres, including aiding replisome assembly genome-wide122, resolving stalled DNA replication forks123,124 and facilitating recovery from DNA damage125–127.
Fig. 3 |. CST and pol α-primase coordination of telomere C-strand fill-in.
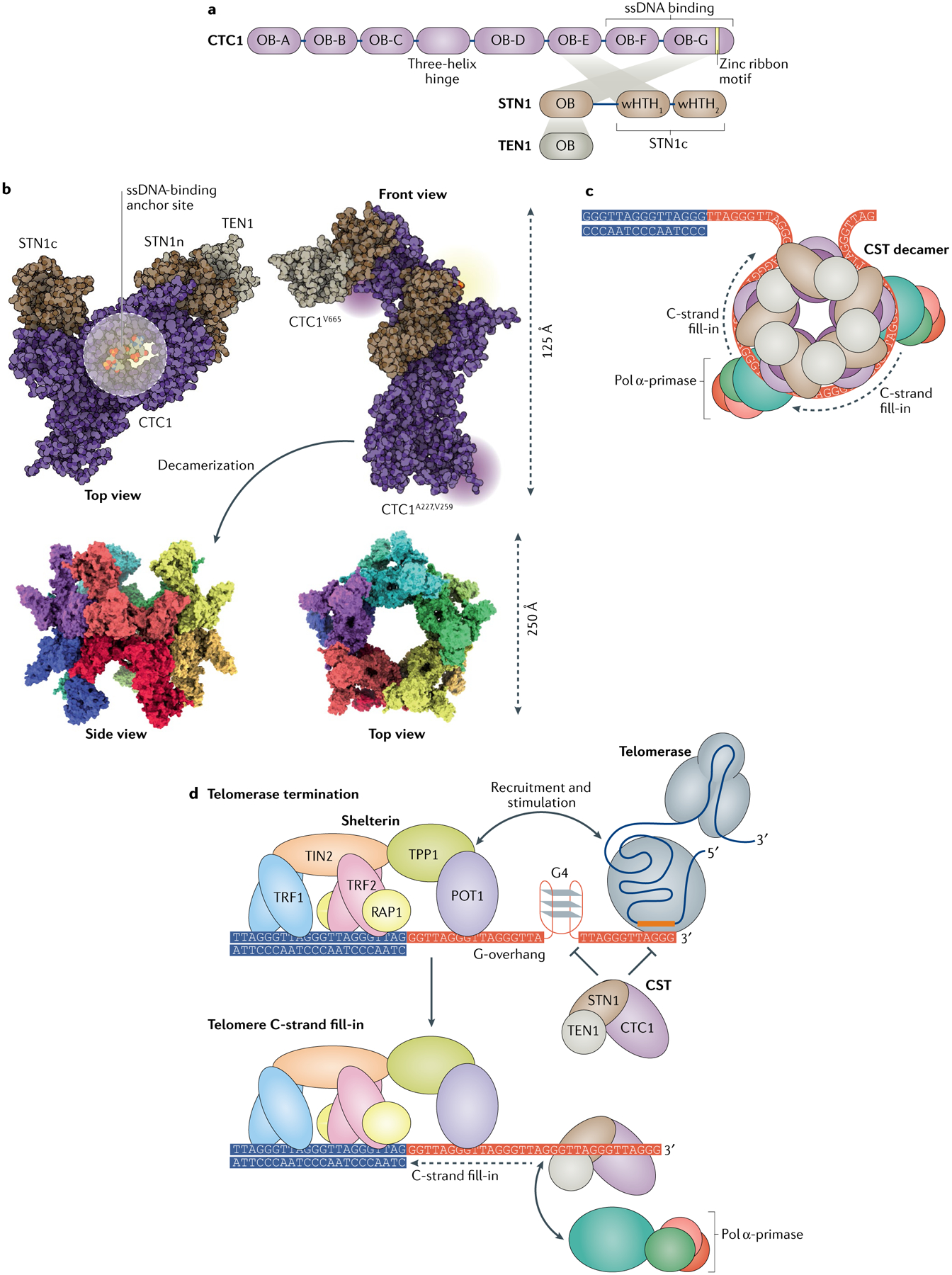
a | Interaction map of human CTC1–STN1–TEN1 (CST) complex subunits and their functional sites. b | Atomic structure of CST, derived from cryo-electron microscopy studies (PDB: 6W6W). The magenta halos on the front view indicate two spatially separated groups of residues known to mediate the interactions of CST with the DNA polymerase α-primase (pol α-primase) complex. CST interaction with telomeric single-stranded DNA (ssDNA) can mediate decamerization of CST. c | CST complex decamerization might enable protection of the telomeric G-overhang tail and loading of multiple pol α-primase proteins at a single G-overhang for facilitation of C-strand fill-in. d | Termination of telomerase activity and C-strand fill-in by shelterin, CST and pol α-primase. The shelterin complex recruits telomerase to telomeres and also stimulates extension of the G-overhang by telomerase. Formation of DNA G-quadruplex (G4) structures could prevent telomerase from engaging the tail, but once telomerase has initiated extension, the G4 structure can aid telomerase translocation. The CST complex might prevent telomerase from engaging the G-overhang through competitive binding, or it might disrupt an ongoing extension process by resolving the G4 structure needed for optimal telomerase translocation. Once the extension process is completed, CST can recruit pol α-primase for lagging strand synthesis of the telomeric C-strand to convert the newly synthesized G-overhang into double-stranded DNA. This process is known as telomere C-strand fill-in. OB, oligonucleotide/oligosaccharide binding; STN1c, STN1 carboxy terminal; STN1n, STN1 amino terminal; POT1, protection of telomeres protein 1; TIN2, TERF1-interacting nuclear factor 2; TRF1, telomeric repeat-binding factor 1; wHTH, winged helix–turn–helix motif.
Replisome.
A multisubunit protein complex that carries out replication of DNA.
Promiscuity of human CST ssDNA binding
ssDNA-binding proteins such as RPA and its bacterial homologue SSB have multiple ssDNA-binding modes128,129. Thus, it is perhaps not surprising that CST, which possesses multiple OB domains, exhibits similar ssDNA-binding behaviour37,38,130–132, giving it the ability to engage a plethora of DNA sequences and structures35,37,133,134. A peculiar finding is that CST binds preferentially to ssDNA–dsDNA junctions, which may explain how CST recognizes specialized DNA structures at DNA replication and breakage sites133. CST binds short ssDNA (<30 nucleotides) with specificity for G-rich sequences, such as telomeric sequences35,134. It can also engage non-telomeric DNA sequences37,133 within longer ssDNA (>30 nucleotides). One model to explain the promiscuous DNA-binding properties of CST postulates that CST has a core site that binds tightly to G-rich ssDNA and also has one or more secondary sites that can accommodate longer ssDNA with less sequence specificity. Indeed, the recent cryo-EM structure of human CST revealed a single telomeric ssDNA-binding site in OB-F and OB-G of the CTC1 subunit, and the presence of five additional OB domains132 (FIG. 3a,b). This model would also explain how CST can engage ssDNA both at telomeres and genome-wide. Other than binding to extended ssDNA structures, CST is also able to directly engage and unfold G4 DNA structures133. This function is crucial for the ability of CST to restore stalled replication forks at telomeres (and genome-wide), presumably by removing the obstacles formed by G4 at the lagging strand124.
Although each subunit of CST has OB domains (which individually interact with ssDNA to some extent), its functional ssDNA-binding capacity depends on its heterotrimeric assembly35,135,136. This could be a functional manifestation of CTC1 topology, whereby STN1–TEN1 stabilizes the CTC1 C-terminus region that contains the core ssDNA-binding site of CST132 (FIG. 3b). This is consistent with previous experiments showing TEN1 stabilizing CTC1–STN1 ssDNA binding137. It is also possible that CST achieves functional ssDNA binding by leveraging multiple cooperative binding sites across the three subunits, similar to RPA138. The current cryo-EM structure of the full CST complex only reveals four nucleotides bound by CTC1, despite the presence of a longer, 18-nucleotide telomeric ssDNA132. CST needs more than four nucleotides to stably engage telomeric ssDNA35, and thus it will be important to determine how and where else CST interacts with ssDNA and how ssDNA wraps around CST. This will not only provide insights into how the heterotrimeric assembly enhances CST affinity for ssDNA37,131 but also how CST can mediate its functions at telomeres and genome-wide at sites of DNA replication and DNA damage repair.
Upon binding to telomeric ssDNA, human CST forms a ring-like decameric DNA–protein supercomplex (FIG. 3b), in which telomeric ssDNA facilitates formation of CST dimers (key building blocks of the decamer)132. Whether the known functions of CST are accomplished by this supercomplex or by CST heterotrimers is currently unknown. However, given the intricate assembly pathway of the decamer and its potential to bury and compact a substantial length of ssDNA, we suggest that decamerization may be involved in DNA protection and in pol α-primase recruitment by CST. For example, a decameric form of CST could recruit multiple pol α-primase proteins (FIG. 3c) to a single telomeric ssDNA site, which would initiate several concurrent and coordinated C-strand fill-in processes, leading to a rapid fill-in of telomeric ssDNA. Loading of multiple DNA polymerases has been observed in the T4 replisome, whereby two DNA polymerases play tag to facilitate a smoother turnover for the lagging-strand DNA synthesis process139. Interestingly, yeast CST also forms a ring-like protein complex, albeit as a dimer, but it is unclear whether ssDNA binding is needed for yeast CST dimerization140.
CST relationship to RPA-like complexes
The human CTC1 subunit has seven OB domains and interacts with STN1 and TEN1 subunits in a 1:1:1 stoichiometry in the heterotrimeric complex132 (FIG. 3b). The stoichiometry is similar to that of human RPA and Tetrahymena thermophila and budding yeast CST103,128,140,141, but not to Candida glabrata CST142. The architecture of human CST indicates that the C-terminal half of CTC1 serves as a structural scaffold for STN1 and TEN1, in which STN1 serves as an adapter between TEN1 and CTC1 (REF.132) (FIG. 3b). Two peculiar findings are that the N-terminal and C-terminal domains of STN1 (STN1n and STN1c, respectively) separately interact with CTC1 and that STN1c can switch between two separate docking sites on CTC1, which is presumably decided by CST binding of ssDNA132. This is the first structural insight into a complete RPA-like complex, and it will be important to determine whether other STN1 homologues (for example, RPA32 or T. thermophila P45) possess this unique architectural assembly, especially as they have very similar structural topology131,141,143.
Unlike STN1–TEN1, CTC1 (the largest subunit in the heterotrimeric complex) is more structurally diverse among RPA-like complexes across species131,132,140,141,144, suggesting that CTC1 has evolved to accommodate various distinct functions. Based on telomere functionality, the budding yeast Cdc13 has often been described as a potential structural homologue of mammalian CTC1, despite the lack of sequence similarity145. The cryo-EM structure of human CTC1 now shows that it resembles the topology of RPA70 more than that of Cdc13 (REFS132,140,144), suggesting that human CTC1 and RPA share a similar evolutionary origin distinct from that of Cdc13. Human CTC1 also contains a conserved zinc ribbon motif in its OB-G domain (FIG. 3a)132, which is similar to that found in RPA and T. thermophila Teb1 (RPA70 homologue). This motif structurally participates in the stimulation of T. thermophila telomerase processivity, although it does not directly bind telomeric ssDNA146. The function of the zinc ribbon motif in CST, as in the case of RPA128, is currently unknown but may involve stimulation of pol α-primase processivity. More studies will be needed to explore the function(s) of the CST zinc ribbon motif, particularly its roles in pol α-primase recruitment and stimulation39,147.
Terminating telomerase activity by CST
A proposed mechanism of termination of telomerase activity is competition between telomerase–TPP1–POT1 and CST for telomeric ssDNA, with CST effectively displacing telomerase from its substrate35 (FIG. 3d). Supporting this model, in vivo studies revealed that a CST disease mutation elevates telomerase recruitment to telomeres136 and that CST recruitment to telomeres coincides with telomerase shut-off at the late S/G2 phase35. As every telomere is likely extended by telomerase only once per cell cycle105,148, it is premature to say whether CST reduces telomerase activity by preventing re-engagement of telomerase to telomeres or by disrupting an elongating telomerase complex. In addition, it is interesting to note that mouse and human use different shelterin-mediated pathways to recruit CST to telomeres: mouse uses POT1b (REF.149) whereas human uses TPP1 (REF.150).
Given that G4 formation at a nascent telomeric tail can aid telomerase processivity116, an interesting question is whether CST unfolding of G4 has any role in the termination of telomerase activity. How CST differs from TPP1–POT1, which can passively disrupt G4 (REF.115), to achieve opposite regulatory effects on telomerase activity is unclear (FIG. 3d). One possibility could be that the actions of TPP1–POT1 and CST are applied at different stages of the telomerase catalytic process. In addition, direct interactions between CST and shelterin35 may also have a role. Further insights might come from understanding how CST and POT1 engage and unfold G4 structures. To reveal such details, it would be important to probe the dynamic interplay between CST, shelterin and telomerase on a single telomeric substrate.
Insights into how human CST interacts with telomerase may come from T. thermophila, where CST is surprisingly an integral part of the telomerase RNP103,145. Furthermore, T. thermophila P50 (a putative TPP1 homologue151) is also part of the telomerase holoenzyme103 and directly interacts with CST (similarly to human TPP1 (REF.35)). Thus, if interactions occurring within a single T. thermophila telomerase holoenzyme are occurring intermolecularly in human telomeres, several insights are revealed. First, analogous CST–TPP1 interactions may contribute to human telomere length regulation35,150,152. Second, the human CST ssDNA-binding site would face towards the 3′ end of the ssDNA, as proposed in the T. thermophila telomerase holoenzyme model103, where it could plausibly bind the product of telomerase extension. In addition, human and T. thermophila CST both have a flexible STN1c domain, which, based on the proposed human CST decamer assembly pathway132, could have a regulatory role in CST interactions with telomeric ssDNA and telomerase. Structures of mammalian telomerase with CST and shelterin bound to a single piece of telomeric ssDNA would test these speculations.
CST coordination of C-strand fill-in
It is often proposed that CST participates in the recruitment of pol α-primase to telomeres, presumably through CST interactions with shelterin or telomeric ssDNA35,120,137,152,153, although this has been challenged121. Even though co-immunoprecipitation studies have pointed to STN1, shelterin proteins and telomerase as potential candidates to recruit pol α-primase to telomeres154, it remains unclear whether these are direct protein–protein interactions or mediated by telomeric DNA associations. A clearer picture exists for pol α-primase recruitment to DNA replication origins, where the replication initiation factors MCM10 and AND-1 mediate its recruitment155,156. Interestingly, recent studies have implicated CST in the assembly of AND-1 and pol α-primase at replication origins, by forming direct interactions with the MCM complex, AND-1 and pol α122. In addition, CST also participates in recruiting pol α-primase to DNA double-strand break sites by interacting with the shieldin complex126,127. All of the above findings support the hypothesis that CST has a direct role in recruiting pol α-primase to telomeres.
Pol α-primase serves two primary functions: the primase catalytic subunit first synthesizes an RNA primer, which is then used by the polymerase catalytic subunit for DNA polymerization. Protein recruitment aside, CST is also involved in stimulating the pol α-primase switch from RNA to DNA synthesis39,157,158. This represents only part of the C-strand fill-in process, the entire mechanism of which remains unclear. Some key questions about C-strand fill-in include how many copies of CST and pol α-primase are present at a single telomeric site, how CST helps primase to initiate RNA synthesis and then switch to DNA synthesis, and whether a G4 formed along the internal telomeric ssDNA will be an obstacle for DNA polymerization by pol α-primase. Answers to these questions would come from structural and biochemical studies of larger assemblies involving CST, pol α-primase, telomerase and shelterin.
Some insights are obtained when CTC1 residues that are involved in the pol α-primase interaction135 are mapped onto its structure, revealing that CST associates with pol α-primase across at least two separate sites132. Interestingly, eukaryotic pol α-primase has a flexible bilobal architecture159, which leads us to speculate that CST could serve as a scaffold to support pol α-primase’s architecture and, possibly, also provide a mechanism to regulate the transfer of RNA primers by modulating the distance between the catalytic and primase subunits of pol α-primase39,158,159.
Disease relevance of shelterin and CST
Shelterin proteins are directly involved in telomere protection and telomerase regulation, and any deviation in their function can lead to abnormal telomere length. Mutations in either telomerase components9 or telomere components160,161 can result in failure to maintain tissues that require constant regeneration, such as skin and bone marrow (see REFS162,163 for selected recent reviews). Furthermore, abnormally short telomeres cause genome instability, which tends to promote cancer. Dyskeratosis congenita is a rare genetic disorder strongly associated with dysfunctional telomere maintenance. More common diseases such as idiopathic pulmonary fibrosis and aplastic anaemia have multiple aetiologies, including telomerase or telomere dysfunction. A comprehensive list164 of telomerase and telomere disease mutations can be found in the Telomerase Database (see Related links). In TABLE 1, we have mapped some of the known disease mutations that reside in structural domains of shelterin proteins and have the possibility to disrupt complex assembly, DNA binding or telomerase recruitment.
TABLE 1 |.
Disease-causing mutations in shelterin and CST proteins and their mapping onto structural domains
| Name of gene (protein) | Affected amino acid | Type of mutation | Structural domains | Associated human diseases | Refs |
|---|---|---|---|---|---|
| TERF1 (TRF1) | 398 | Y to C | Myb (DNA binding)b | Melanoma | 194 |
| TERF2 (TRF2)a | 183 | T to M | TRFHc | Hepatocellular carcinoma | 195 |
| 265 | R to H | TRFHc | Melanoma | 194 | |
| 498 | E to K | Mybb | Breast cancer | 194,196 | |
| TERF2IP (RAP1) | 364 | R to Δ | TRF2-binding domainc | Melanoma | 197 |
| TINF2 (TIN2) | 269 | E to Δ | TRF1-binding domainc | Bone marrow failure, dyskeratosis congenita | 198,199 |
| 271 | E to Δ | Aplastic anaemia | 200 | ||
| 276 | R to G | Bone marrow failure | 198 | ||
| ACD (TPP1)a | 84 | K to Δ | OB domain (TEL patch)d | Dyskeratosis congenita | 165,166 |
| 114 | A to T | Melanoma | 197 | ||
| 163 | N to S | ||||
| 186 | V to M | POT1-binding domainc | |||
| 234 | Q to Δ | ||||
| 236 | I to F | ||||
| POT1 (POT1) | 89 | Y to C | OB domainse | Melanoma | 201 |
| 94 | Q to E | ||||
| 273 | R to L | ||||
| 223 | Y to C | ||||
| 446 | P to Q | TPP1-binding domainc | Chronic lymphocytic leukaemia | 60 | |
| 591 | C to W | ||||
| 623 | Q to H | ||||
| CTC1 (CTC1) | 227 | A to V | OB-B domainf | Cerebroretinal microangiopathy | 132,135,202 |
| 259 | V to M | Coats plus | 132,135,168 | ||
| 484 | H to P | Hinge three-helix bundleg | Coats plus | 132,203 | |
| 665 | V to G | OB-D domainf | Cerebroretinal microangiopathy | 132,135,202 | |
| 840 | R to W | OB-E domaine | Coats plus | 132,135,168 | |
| 871 | V to M | OB-F domaine | Coats plus | 132,135,168 | |
| 975 | R to G | OB-F domaine | Cerebroretinal microangiopathy, Coats plus | 132,135,168,202 | |
| 987 | R to W | OB-F domaine | Coats plus, dyskeratosis congenita | 132,135,168,204 | |
| 1195 | R to Δ | OB-G domainh | Cerebroretinal microangiopathy, Coats plus | 132,135,168,202 | |
| STN1 (STN1) | 135 | R to T | TEN1 and CTC1-binding domainh | Coats plus | 132,169 |
Unless otherwise stated, data are compiled from the Telomerase Database and the Human Gene Mutation Database.
CST, CTC1–STN1–TEN1; OB, oligonucleotide/oligosaccharide binding; POT1, protection of telomeres protein 1; TEL patch, TPP1 glutamate (E) and leucine (L)-rich patch; TIN2, TERF1-interacting nuclear factor 2; TRF1, telomeric repeat-binding factor 1; TRFH, TRF homology domain.
Amino acids of TRF2 and TPP1 are annotated as in NP_005643.2 and NP_001075955.2, respectively.
Involved in binding of telomeric double-stranded DNA.
Involved in shelterin assembly.
Involved in telomerase recruitment.
Involved in binding of telomeric single-stranded DNA.
Mutation disrupts interaction with DNA polymerase α-primase.
Mutation disrupts CST decamer assembly.
Involved in CST complex formation.
An area of extensive structure–function relationship studies is the recruitment of telomerase by the TEL patch of the OB domain of TPP1 (REFS32–34). These findings have been used to relate a mutation (ΔK170) in the TEL patch, associated with an inherited bone marrow failure, to a telomerase recruitment defect165. Further studies have revealed that the TPP1 ΔK170 mutation results in a major restructuring of the TEL patch region, causing the loss of interaction between TPP1 and telomerase166.
The conserved CST complex participates in multiple processes that are crucial for genome stability. CST dysfunction can lead to the deregulation of telomere length, replication fork stalling and deregulation of DNA damage repair, all of which lead to genome instability. The common manifestations of CST dysfunction are Coats plus syndrome and dyskeratosis congenita167,168. These diseases stem from mutations in CTC1 and STN1 (REFS167–169) that cause various loss-of-function consequences in CST complex assembly, ssDNA binding and pol α-primase association135,170,171. Known disease mutations are mapped onto the structural domains of human CST in TABLE 1.
Now that most of the key interactions involved in shelterin, CST and telomerase assembly have been determined, we expect future state-of-the-art multi-omics analyses to reveal additional relationships between deregulation of telomeric structure and human disease.
Other human telomere proteins
In this Review, we focus on the shelterin and CST complexes, because there have been recent breakthroughs in understanding their structure–function relationships. In this section, we briefly discuss a few of the many other proteins that bind directly to telomeric DNA. We do not cover the signalling complexes that act at telomeres, as these have been recently reviewed172.
The other prominent telomeric dsDNA-binding proteins are nucleosomal histone proteins. The nucleosomal architecture of mammalian telomeric chromatin was shown early to consist of highly organized nucleosome arrays with very short (10-bp) linkers between nucleosomes21,173–175. A simple model would posit that these nucleosome arrays occupy the inner portions of telomeres and that the shelterin complexes coat nucleosome-free DNA closer to the ssDNA tail, but this is not the case. Instead, the ordered nucleosome arrays continue all the way to the tail175. Furthermore, TRF1 can bind to the DNA gyre of nucleosomes176. By contrast, TRF2 binding is inhibited by nucleosomes (and vice versa177), so the linker DNA may be particularly important for TRF2 binding. Perhaps the more dynamic and thermodynamically less stable nucleosomes formed with TTAGGG repeat sequences178,179 facilitate TRF2 binding. In the future, structures of telomeric nucleosome–shelterin complexes should provide insight into how shelterin and nucleosomes cohabit at telomeres.
The heterochromatin nature of mammalian telomeres was presaged by cytological studies in fruit flies180 and by analysis of the telomere position effect (epigenetic repression of transcription) in yeast181. Human telomeric chromatin is enriched in histone H3 trimethylated at Lys9 (H3K9me3), which is a mark of constitutive heterochromatin. H3K9me3 is bound by heterochromatin protein 1 (HP1), and HP1 dysfunction leads to abnormal telomere elongation182 and deregulation of TERRA transcription183. Unexpectedly, H3K27me3 of facultative heterochromatin coexists with H3K9me3 at telomeres183. Loss of these repressive histone modifications has consequences for telomere length maintenance, likely owing to the deregulation of protein accessibility to telomeres caused by chromatin structural changes184.
Unbiased proteomic analyses of telomeric proteins revealed about 200 proteins185,186. In addition to the canonical histones, the histone variants macroH2A and testis-specific H2B were prominent at telomeres185. Unanticipated members of the telomere proteome included all subunits of the THO complex186. THO counteracts telomeric R-loops formed by invasion of TERRA RNA into telomeric DNA, thereby facilitating telomere replication and length maintenance187.
The Ku heterodimer is required for the repair of DNA double-strand breaks by non-homologous DNA end-joining. Thus, it is paradoxical that Ku is found at telomeres, which must not undergo end-joining. It has been proposed that the ability of Ku to initiate non-homologous DNA end-joining at telomeric DNA ends is blocked by its interaction with TRF2 (REF.188). Telomeric functions of Ku have been most extensively studied in yeast189, and it is not clear how many of these functions occur in mammals. One function of mammalian Ku is to protect telomeres from homologous recombination190.
Telomeric zinc finger-associated protein (TZAP) binds preferentially to telomeric sequences in competition with TRF1 and TRF2 (REF.24). Because TZAP binds preferentially to long telomeres and initiates telomere trimming, it appears to help control the upper limit of telomere length. How TZAP coordinates its function with shelterin and their respective distributions along telomeres of various lengths remain open questions.
Considering proteins with affinity to ssDNA, hnRNP proteins, including hnRNPA1 and hnRNPA2/B1, have specificity for the telomeric sequence. Although these hnRNP proteins bind with higher affinity to RNA than to DNA, they are highly abundant in nuclei and therefore bind to telomeric ssDNA as well191,192. Mouse cells depleted of hnRNPA1 have short telomeres193. hnRNPA1 also displaces RPA, but not POT1, from telomeric ssDNA, an activity that is suppressed during telomere replication through sequestration of hnRNPA1 by TERRA111.
The structure of an entire telomere?
Like snowflakes, no two telomeres are identical. But how can we make such a bold claim? First, even within a single nucleus, telomeres differ in length, so they cannot have the same structure in detail. Second, even two telomeres of identical length are highly unlikely to have their sequence-specific DNA-binding proteins residing at the same telomeric repeat positions; this would require unprecedented and almost inconceivable regulatory processes to achieve such uniformity, with doubtful selective advantage. Absence of uniformity is seen also at T-loops, where the loop size varies from small to encompassing, essentially, the entire telomeric DNA19; a single telomere with exactly 12 kb of dsDNA (2,000 telomeric repeats) can presumably form almost 2,000 different lengths of T-loops, with very small loops being disfavoured by the inherent stiffness of dsDNA. Finally, telomere binding by proteins and RNPs is dynamic, so any given telomere structure will change rapidly over time.
Thus, when we speak of ‘telomere structure’, we are always speaking of an ensemble of related structures. One can view telomere regulation at two different scales (FIG. 4a): the higher-order chromatin architecture that affects telomere accessibility and the hive of localized telomerase-related interactions at the telomere 3′ tail (the ‘telomere replisome’). For both of these regimes, we strive to know the atomic-resolution structures of their elemental units, including shelterin with and without the nucleosome, CST and telomerase, and to know how these units are arrayed along the telomere, for example, how far from the dsDNA–ssDNA junction shelterin complexes are located, whether there is some order to their distribution or they are randomly spaced and how the array changes with T-loop formation or cell cycle progression. We also need to determine how the switches between different structural states of telomeres are orchestrated and regulated (FIG. 4b), and the dynamics of key binding events and functional interactions. At the same time, we need to address the assembly mechanism (FIG. 4c), for example, whether shelterin components slide or hop along the DNA or partake in a 3D diffusional search.
Fig. 4 |. Assembling the telomeres.
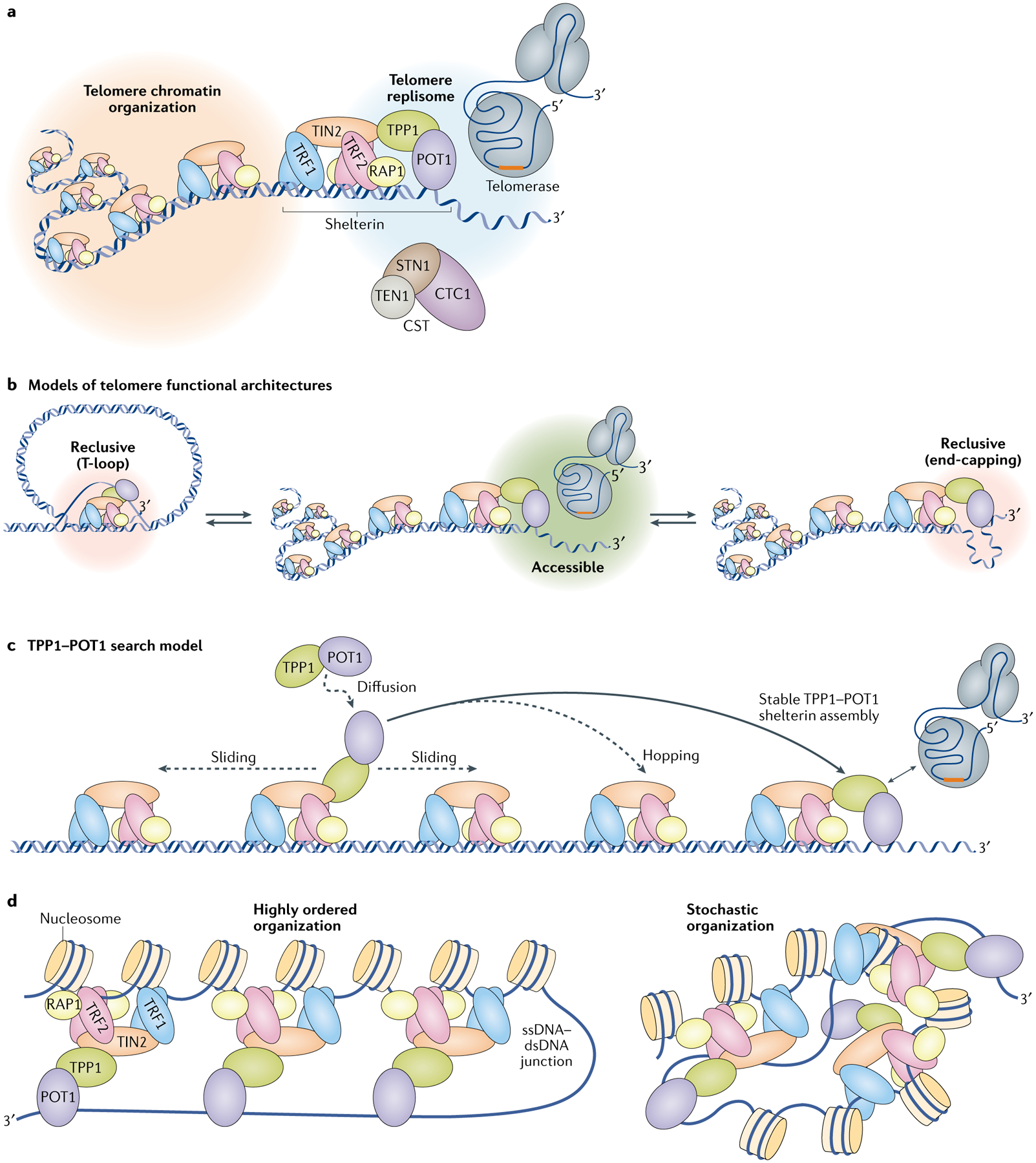
a | Telomere maintenance is regulated at two genomic scales: modulation of higher-order DNA–protein architecture across the telomere (gold cloud) and regulation of telomerase activity at the G-overhang (the ‘telomere replisome’; blue cloud). b | The telomeric 3′ tail can be made accessible (green cloud) or reclusive (red clouds) by telomeric protein complexes. The shelterin complex facilitates telomere loop (T-loop) formation, which restricts telomerase access by burying the 3′ tail in double-stranded telomeric DNA. A T-loop can unravel to a linear form to allow telomerase access, which can be further regulated through end-capping by telomeric proteins. c | How TPP1–protection of telomeres protein 1 (POT1) might find the G-overhang among multiple similar binding sites once the tail is made accessible. For simplicity, nucleosomes and other telomeric proteins are omitted from the cartoon. Possible mechanisms of TPP1–POT1 delivery to the G-overhang include 3D diffusion, sliding or hopping along telomeres. Solid and dashed lines indicate stable and transient interactions, respectively. d | Illustrations of two possible and polar opposite outcomes of telomere organization driven by shelterin complexes. A highly ordered shelterin array could result in a zipper-like folding of the telomere. On the other hand, a random deposition of shelterin would result in a disordered telomere architecture. Either possibility restricts access to the telomere 3′ tail and also leads to higher-order telomere organization. CST, CTC1–STN1–TEN1; dsDNA, double-stranded DNA; ssDNA, single-stranded DNA; TIN2, TERF1-interacting nuclear factor 2; TRF1, telomeric repeat-binding factor 1.
Experiments addressing these questions are challenged by the repetitive sequence of the telomere. Thus, techniques that are powerful for studying unique sequences, such as chromatin immunoprecipitation (ChIP) and ChIP followed by sequencing (ChIP–seq), can reveal that a protein is telomeric but not where along a telomere it is located. To illustrate the primitive state of our current knowledge, we have not yet determined whether entire telomeres are highly ordered or randomly organized (FIG. 4d), or represent something in between.
Conclusions and future perspective
Recent years have seen telomere biology research greatly benefitting from applications of cutting-edge techniques. Individual endogenous telomerase RNPs can be tracked in living human cells and their interactions with telomeres can be interrogated105–107. It is now important to extend similar approaches to other telomeric protein complexes, such as shelterin, CST and TZAP, which would be especially informative in combination with multicolour single-molecule imaging. Before the cryo-EM revolution, telomere structural biology had been relatively stagnant for a period of time. Because telomerase and telomeres are large and some-what flexible protein–nucleic acid complexes, numerous years of effort by X-ray crystallography and nuclear magnetic resonance approaches had been restricted to analysis of stable domains. Now, cryo-EM analysis has allowed the elucidation of the elusive architectures of T. thermophila and human telomerases103,104 and the atomic structure of human CST132. As the technology of cryo-EM analysis improves and becomes amenable to more flexible macromolecular structures, determining the architectures of several important telomeric protein–nucleic acid complexes — mammalian shelterin with telomeric DNA, or telomerase with shelterin and/or CST, or CST–pol α-primase — will be of high priority.
Thus, the next key steps to understanding the molecular details of elemental telomeric components are reasonably clear. However, a grander goal exists. As discussed above, determining the structure of an entire telomere is not straightforward and likely presents a more complicated pathway forging forward. This is because new technology will be needed to breach the repetitive nature of telomeric DNA sequences, ideally one that would allow us to harness our current familiarity with next-generation sequencing techniques and bioinformatics analysis. Emerging techniques in super-resolution 3D and 4D chromatin imaging or in cryo-electron tomography could also help unveil native telomere structures in cells. Thus, with a clever combination of techniques, we may well reveal the heterogeneous landscape of telomere flora and fauna across both space and time.
Acknowledgements
The authors thank T. de Lange for helpful comments and suggestions. C.L. thanks the members of the Cech laboratory for their support and advice during his postdoctoral years at the University of Colorado Boulder. This work was funded in part by the National Institutes of Health (NIH; R00GM131023) and the Steenbock Career Award from the University of Wisconsin-Madison to C.L. T.R.C. is an investigator at the Howard Hughes Medical Institute.
Footnotes
Competing interests
T.R.C. is on the board of directors of Merck and a consultant for STORM Therapeutics and Eikon Therapeutics.
RELATED LINKS
Human Gene Mutation Database: http://www.hgmd.cf.ac.uk/
Telomerase Database: http://telomerase.asu.edu
References
- 1.Moyzis RK et al. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl Acad. Sci. USA 85, 6622–6626 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyne J, Ratliff RL & Moyzis RK Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc. Natl Acad. Sci. USA 86, 7049–7053 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Makarov VL, Hirose Y & Langmore JP Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell 88, 657–666 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Lansdorp PM et al. Heterogeneity in telomere length of human chromosomes. Hum. Mol. Genet 5, 685–691 (1996). [DOI] [PubMed] [Google Scholar]
- 5.Vaziri H et al. Loss of telomeric DNA during aging of normal and trisomy 21 human lymphocytes. Am. J. Hum. Genet 52, 661–667 (1993). [PMC free article] [PubMed] [Google Scholar]
- 6.Allsopp RC et al. Telomere length predicts replicative capacity of human fibroblasts. Proc. Natl Acad. Sci. USA 89, 10114–10118 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harley CB, Futcher AB & Greider CW Telomeres shorten during ageing of human fibroblasts. Nature 345, 458–460 (1990). [DOI] [PubMed] [Google Scholar]
- 8.Herbig U, Jobling WA, Chen BP, Chen DJ & Sedivy JM Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21CIP1, but not p16INK4a. Mol. Cell 14, 501–513 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Mitchell JR, Wood E & Collins K A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402, 551–555 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Vulliamy TJ, Knight SW, Mason PJ & Dokal I Very short telomeres in the peripheral blood of patients with X-linked and autosomal dyskeratosis congenita. Blood Cell Mol. Dis 27, 353–357 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Kim NW et al. Specific association of human telomerase activity with immortal cells and cancer. Science 266, 2011–2015 (1994). [DOI] [PubMed] [Google Scholar]
- 12.Shay JW & Bacchetti S A survey of telomerase activity in human cancer. Eur. J. Cancer 33, 787–791 (1997). [DOI] [PubMed] [Google Scholar]
- 13.Ye JZ et al. TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J. Biol. Chem 279, 47264–47271 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Liu D, O’Connor MS, Qin J & Songyang Z Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J. Biol. Chem 279, 51338–51342 (2004). [DOI] [PubMed] [Google Scholar]
- 15.O’Connor MS, Safari A, Xin H, Liu D & Songyang Z A critical role for TPP1 and TIN2 interaction in high-order telomeric complex assembly. Proc. Natl Acad. Sci. USA 103, 11874–11879 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]; Together with Liu et al. (J. Biol. Chem. 2004), this study identifies the full six-member shelterin complex, showing TIN2 binding both TRF1 and TRF2.
- 16.de Lange T Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19, 2100–2110 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Baumann P & Cech TR Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292, 1171–1175 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Lei M, Podell ER & Cech TR Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol 11, 1223–1229 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Griffith JD et al. Mammalian telomeres end in a large duplex loop. Cell 97, 503–514 (1999). [DOI] [PubMed] [Google Scholar]; This study reports that a substantial fraction of mammalian telomeres form a dsDNA loop to protect chromosome ends.
- 20.Stansel RM, de Lange T & Griffith JD T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J 20, 5532–5540 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nikitina T & Woodcock CL Closed chromatin loops at the ends of chromosomes. J. Cell Biol 166, 161–165 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doksani Y, Wu JY, de Lange T & Zhuang X Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell 155, 345–356 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marcand S, Gilson E & Shore D A protein-counting mechanism for telomere length regulation in yeast. Science 275, 986–990 (1997). [DOI] [PubMed] [Google Scholar]
- 24.Li JS et al. TZAP: a telomere-associated protein involved in telomere length control. Science 355, 638–641 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study reports a telomeric DNA-binding protein that initiates trimming of long telomeres.
- 25.Greider CW & Blackburn EH A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337, 331–337 (1989). [DOI] [PubMed] [Google Scholar]
- 26.Lingner J et al. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276, 561–567 (1997). [DOI] [PubMed] [Google Scholar]; This study and Greider and Blackburn (1989) report the first cloning of genes for the RNA and the catalytic (TERT) subunits of telomerase in ciliated protozoa, which led to the identification of the corresponding human factors.
- 27.Greider CW Telomerase is processive. Mol. Cell Biol 11, 4572–4580 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roake CM & Artandi SE Regulation of human telomerase in homeostasis and disease. Nat. Rev. Mol. Cell Biol 21, 384–397 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt JC & Cech TR Human telomerase: biogenesis, trafficking, recruitment, and activation. Genes Dev 29, 1095–1105 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hockemeyer D & Collins K Control of telomerase action at human telomeres. Nat. Struct. Mol. Biol 22, 848–852 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wright WE, Piatyszek MA, Rainey WE, Byrd W & Shay JW Telomerase activity in human germline and embryonic tissues and cells. Dev. Genet 18, 173–179 (1996). [DOI] [PubMed] [Google Scholar]
- 32.Nandakumar J et al. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature 492, 285–289 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong FL et al. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell 150, 481–494 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sexton AN, Youmans DT & Collins K Specificity requirements for human telomere protein interaction with telomerase holoenzyme. J. Biol. Chem 287, 34455–34464 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]; Together with Nandakumar et. al. (2012) and Zhong et. al. (2012), this study identifies specific amino acids of the TPP1 OB fold that recruit telomerase and enhance its enzymatic processivity.
- 35.Chen LY, Redon S & Lingner J The human CST complex is a terminator of telomerase activity. Nature 488, 540–544 (2012). [DOI] [PubMed] [Google Scholar]
- 36.Gao H, Cervantes RB, Mandell EK, Otero JH & Lundblad V RPA-like proteins mediate yeast telomere function. Nat. Struct. Mol. Biol 14, 208–214 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Miyake Y et al. RPA-like mammalian Ctc1–Stn1–Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol. Cell 36, 193–206 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Surovtseva YV et al. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol. Cell 36, 207–218 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]; Together with Miyake et al. (2009), this study shows that the RPA-like CST complex, previously characterized in yeast telomeres, has mammalian and plant counterparts with telomeric functions.
- 39.Casteel DE et al. A DNA polymerase-α primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J. Biol. Chem 284, 5807–5818 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E & Lingner J Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318, 798–801 (2007). [DOI] [PubMed] [Google Scholar]
- 41.van Steensel B, Smogorzewska A & de Lange T TRF2 protects human telomeres from end-to-end fusions. Cell 92, 401–413 (1998). [DOI] [PubMed] [Google Scholar]; This study shows that interfering with TRF2 accumulation at telomeres causes dramatic chromosome end fusions.
- 42.Smogorzewska A et al. Control of human telomere length by TRF1 and TRF2. Mol. Cell Biol 20, 1659–1668 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim SH, Kaminker P & Campisi J TIN2, a new regulator of telomere length in human cells. Nat. Genet 23, 405–412 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li B & de Lange T Rap1 affects the length and heterogeneity of human telomeres. Mol. Biol. Cell 14, 5060–5068 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye JZ et al. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev 18, 1649–1654 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu D et al. PTOP interacts with POT1 and regulates its localization to telomeres. Nat. Cell Biol 6, 673–680 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Colgin LM, Baran K, Baumann P, Cech TR & Reddel RR Human POT1 facilitates telomere elongation by telomerase. Curr. Biol 13, 942–946 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Bianchi A, Smith S, Chong L, Elias P & de Lange T TRF1 is a dimer and bends telomeric DNA. EMBO J 16, 1785–1794 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Broccoli D, Smogorzewska A, Chong L & de Lange T Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet 17, 231–235 (1997). [DOI] [PubMed] [Google Scholar]
- 50.van Steensel B & de Lange T Control of telomere length by the human telomeric protein TRF1. Nature 385, 740–743 (1997). [DOI] [PubMed] [Google Scholar]
- 51.Bilaud T et al. Telomeric localization of TRF2, a novel human telobox protein. Nat. Genet 17, 236–239 (1997). [DOI] [PubMed] [Google Scholar]
- 52.Loayza D & De Lange T POT1 as a terminal transducer of TRF1 telomere length control. Nature 423, 1013–1018 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Lim C, Zaug AJ, Kim HJ & Cech TR Reconstitution of human shelterin complexes reveals unexpected stoichiometry and dual pathways to enhance telomerase processivity. Nat. Commun 8, 1075 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rai R, Chen Y, Lei M & Chang S TRF2–RAP1 is required to protect telomeres from engaging in homologous recombination-mediated deletions and fusions. Nat. Commun 7, 10881 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sfeir A, Kabir S, van Overbeek M, Celli GB & de Lange T Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science 327, 1657–1661 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lototska L et al. Human RAP1 specifically protects telomeres of senescent cells from DNA damage. EMBO Rep 21, e49076 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang F et al. The POT1–TPP1 telomere complex is a telomerase processivity factor. Nature 445, 506–510 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Xin H et al. TPP1 is a homologue of ciliate TEBP-β and interacts with POT1 to recruit telomerase. Nature 445, 559–562 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Lei M, Zaug AJ, Podell ER & Cech TR Switching human telomerase on and off with hPOT1 protein in vitro. J. Biol. Chem 280, 20449–20456 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Rice C et al. Structural and functional analysis of the human POT1–TPP1 telomeric complex. Nat. Commun 8, 14928 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen C et al. Structural insights into POT1–TPP1 interaction and POT1 C-terminal mutations in human cancer. Nat. Commun 8, 14929 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kar A, Willcox S & Griffith JD Transcription of telomeric DNA leads to high levels of homologous recombination and T-loops. Nucleic Acids Res 44, 9369–9380 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu C et al. Structural and functional analyses of the mammalian TIN2–TPP1–TRF2 telomeric complex. Cell Res 27, 1485–1502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study reports the crystal structure of the key ‘bridge’ region of mammalian shelterin, which connects its dsDNA-binding and ssDNA-binding subunits.
- 64.Chen LY et al. Mitochondrial localization of telomeric protein TIN2 links telomere regulation to metabolic control. Mol. Cell 47, 839–850 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen Y et al. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science 319, 1092–1096 (2008). [DOI] [PubMed] [Google Scholar]
- 66.Fairall L, Chapman L, Moss H, de Lange T & Rhodes D Structure of the TRFH dimerization domain of the human telomeric proteins TRF1 and TRF2. Mol. Cell 8, 351–361 (2001). [DOI] [PubMed] [Google Scholar]
- 67.Bianchi A et al. TRF1 binds a bipartite telomeric site with extreme spatial flexibility. EMBO J 18, 5735–5744 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nishikawa T, Nagadoi A, Yoshimura S, Aimoto S & Nishimura Y Solution structure of the DNA-binding domain of human telomeric protein, hTRF1. Structure 6, 1057–1065 (1998). [DOI] [PubMed] [Google Scholar]
- 69.Court R, Chapman L, Fairall L & Rhodes D How the human telomeric proteins TRF1 and TRF2 recognize telomeric DNA: a view from high-resolution crystal structures. EMBO Rep 6, 39–45 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smith S & de Lange T TRF1, a mammalian telomeric protein. Trends Genet 13, 21–26 (1997). [DOI] [PubMed] [Google Scholar]
- 71.Griffith J, Bianchi A & de Lange T TRF1 promotes parallel pairing of telomeric tracts in vitro. J. Mol. Biol 278, 79–88 (1998). [DOI] [PubMed] [Google Scholar]
- 72.Seimiya H, Muramatsu Y, Smith S & Tsuruo T Functional subdomain in the ankyrin domain of tankyrase 1 required for poly(ADP-ribosyl)ation of TRF1 and telomere elongation. Mol. Cell Biol 24, 1944–1955 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Poulet A et al. The N-terminal domains of TRF1 and TRF2 regulate their ability to condense telomeric DNA. Nucleic Acids Res 40, 2566–2576 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Saint-Leger A et al. The basic N-terminal domain of TRF2 limits recombination endonuclease action at human telomeres. Cell Cycle 13, 2469–2474 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Necasova I, Janouskova E, Klumpler T & Hofr C Basic domain of telomere guardian TRF2 reduces D-loop unwinding whereas Rap1 restores it. Nucleic Acids Res 45, 12599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schmutz I, Timashev L, Xie W, Patel DJ & de Lange T TRF2 binds branched DNA to safeguard telomere integrity. Nat. Struct. Mol. Biol 24, 734–742 (2017). [DOI] [PubMed] [Google Scholar]
- 77.Konishi A, Izumi T & Shimizu S TRF2 protein interacts with core histones to stabilize chromosome ends. J. Biol. Chem 291, 20798–20810 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang RC, Smogorzewska A & de Lange T Homologous recombination generates T-loop-sized deletions at human telomeres. Cell 119, 355–368 (2004). [DOI] [PubMed] [Google Scholar]
- 79.Fouche N et al. The basic domain of TRF2 directs binding to DNA junctions irrespective of the presence of TTAGGG repeats. J. Biol. Chem 281, 37486–37495 (2006). [DOI] [PubMed] [Google Scholar]
- 80.Williamson JR, Raghuraman MK & Cech TR Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell 59, 871–880 (1989). [DOI] [PubMed] [Google Scholar]
- 81.Sundquist WI & Klug A Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature 342, 825–829 (1989). [DOI] [PubMed] [Google Scholar]
- 82.Pedroso IM, Hayward W & Fletcher TM The effect of the TRF2 N-terminal and TRFH regions on telomeric G-quadruplex structures. Nucleic Acids Res 37, 1541–1554 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Janouskova E et al. Human Rap1 modulates TRF2 attraction to telomeric DNA. Nucleic Acids Res 43, 2691–2700 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Choi KH, Farrell AS, Lakamp AS & Ouellette MM Characterization of the DNA binding specificity of Shelterin complexes. Nucleic Acids Res 39, 9206–9223 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Erdel F et al. Telomere recognition and assembly mechanism of mammalian shelterin. Cell Rep 18, 41–53 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hanaoka S, Nagadoi A & Nishimura Y Comparison between TRF2 and TRF1 of their telomeric DNA-bound structures and DNA-binding activities. Protein Sci 14, 119–130 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nishikawa T et al. Structure of the DNA-binding domain of human telomeric protein, TRF1 and its interaction with telomeric DNA. Nucleic Acids Res. Suppl 1, 273–274 (2001). [DOI] [PubMed] [Google Scholar]
- 88.Takai KK, Hooper S, Blackwood S, Gandhi R & de Lange T In vivo stoichiometry of shelterin components. J. Biol. Chem 285, 1457–1467 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pike AM, Strong MA, Ouyang JPT & Greider CW TIN2 functions with TPP1/POT1 to stimulate telomerase processivity. Mol. Cell Biol 39, e00593–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nelson ND et al. The C-terminal extension unique to the long isoform of the shelterin component TIN2 enhances its interaction with TRF2 in a phosphorylation- and dyskeratosis congenita cluster-dependent fashion. Mol. Cell Biol 38, e00025–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Grill S et al. Two separation-of-function isoforms of human TPP1 dictate telomerase regulation in somatic and germ cells. Cell Rep 27, 3511–3521.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zemp I & Lingner J The shelterin component TPP1 is a binding partner and substrate for the deubiquitinating enzyme USP7. J. Biol. Chem 289, 28595–28606 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bhanot M & Smith S TIN2 stability is regulated by the E3 ligase Siah2. Mol. Cell Biol 32, 376–384 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Janovic T, Stojaspal M, Veverka P, Horakova D & Hofr C Human telomere repeat binding factor TRF1 replaces TRF2 bound to shelterin core hub TIN2 when TPP1 is absent. J. Mol. Biol 431, 3289–3301 (2019). [DOI] [PubMed] [Google Scholar]
- 95.Kim JK et al. Structural basis for shelterin bridge assembly. Mol. Cell 68, 698–714.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang Y et al. Phosphorylation of TPP1 regulates cell cycle-dependent telomerase recruitment. Proc. Natl Acad. Sci. USA 110, 5457–5462 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gray JT, Celander DW, Price CM & Cech TR Cloning and expression of genes for the Oxytricha telomere-binding protein: specific subunit interactions in the telomeric complex. Cell 67, 807–814 (1991). [DOI] [PubMed] [Google Scholar]
- 98.Yang Q, Zheng YL & Harris CC POT1 and TRF2 cooperate to maintain telomeric integrity. Mol. Cell Biol 25, 1070–1080 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Timashev LA & de Lange T Characterization of T-loop formation by TRF2. Nucleus 11, 164–177 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Amiard S et al. A topological mechanism for TRF2-enhanced strand invasion. Nat. Struct. Mol. Biol 14, 147–154 (2007). [DOI] [PubMed] [Google Scholar]
- 101.Subramaniam S The cryo-EM revolution: fueling the next phase. IUCrJ 6, 1–2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cheng Y Single-particle cryo-EM — how did it get here and where will it go. Science 361, 876–880 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jiang J et al. Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions. Science 350, aab4070 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nguyen THD et al. Cryo-EM structure of substrate-bound human telomerase holoenzyme. Nature 557, 190–195 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Schmidt JC, Zaug AJ & Cech TR Live cell imaging reveals the dynamics of telomerase recruitment to telomeres. Cell 166, 1188–1197.e9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schmidt JC, Zaug AJ, Kufer R & Cech TR Dynamics of human telomerase recruitment depend on template-telomere base pairing. Mol. Biol. Cell 29, 869–880 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Laprade H et al. Single-molecule imaging of telomerase RNA reveals a recruitment-retention model for telomere elongation. Mol. Cell 79, 115–126.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 108.Lin J et al. TRF1 and TRF2 use different mechanisms to find telomeric DNA but share a novel mechanism to search for protein partners at telomeres. Nucleic Acids Res 42, 2493–2504 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hockemeyer D, Sfeir AJ, Shay JW, Wright WE & de Lange T POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J 24, 2667–2678 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Denchi EL & de Lange T Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448, 1068–1071 (2007). [DOI] [PubMed] [Google Scholar]; This study establishes roles of shelterin components in subduing specific signalling pathways that detect DNA damage but need to be repressed at natural chromosome ends.
- 111.Flynn RL et al. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature 471, 532–536 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang H, Nora GJ, Ghodke H & Opresko PL Single molecule studies of physiologically relevant telomeric tails reveal POT1 mechanism for promoting G-quadruplex unfolding. J. Biol. Chem 286, 7479–7489 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zaug AJ, Podell ER & Cech TR Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc. Natl Acad. Sci. USA 102, 10864–10869 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zahler AM, Williamson JR, Cech TR & Prescott DM Inhibition of telomerase by G-quartet DNA structures. Nature 350, 718–720 (1991). [DOI] [PubMed] [Google Scholar]
- 115.Hwang H, Buncher N, Opresko PL & Myong S POT1–TPP1 regulates telomeric overhang structural dynamics. Structure 20, 1872–1880 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jansson LI et al. Telomere DNA G-quadruplex folding within actively extending human telomerase. Proc. Natl Acad. Sci. USA 116, 9350–9359 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Patrick EM, Slivka JD, Payne B, Comstock MJ & Schmidt JC Observation of processive telomerase catalysis using high-resolution optical tweezers. Nat. Chem. Biol 16, 801–809 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Latrick CM & Cech TR POT1–TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J 29, 924–933 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Paudel BP et al. A mechanism for the extension and unfolding of parallel telomeric G-quadruplexes by human telomerase at single-molecule resolution. eLife 9, e56428 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang F et al. Human CST has independent functions during telomere duplex replication and C-strand fill-in. Cell Rep 2, 1096–1103 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Huang C, Dai X & Chai W Human Stn1 protects telomere integrity by promoting efficient lagging-strand synthesis at telomeres and mediating C-strand fill-in. Cell Res 22, 1681–1695 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang Y, Brady KS, Caiello BP, Ackerson SM & Stewart JA Human CST suppresses origin licensing and promotes AND-1/Ctf4 chromatin association. Life Sci. Alliance 2, e201800270 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stewart JA et al. Human CST promotes telomere duplex replication and general replication restart after fork stalling. EMBO J 31, 3537–3549 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang M et al. Mammalian CST averts replication failure by preventing G-quadruplex accumulation. Nucleic Acids Res 47, 5243–5259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang F, Stewart J & Price CM Human CST abundance determines recovery from diverse forms of DNA damage and replication stress. Cell Cycle 13, 3488–3498 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Barazas M et al. The CST complex mediates end protection at double-strand breaks and promotes PARP inhibitor sensitivity in BRCA1-deficient cells. Cell Rep 23, 2107–2118 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mirman Z et al. 53BP1-RIF1-shieldin counteracts DSB resection through CST- and Polα-dependent fill-in. Nature 560, 112–116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bochkareva E, Korolev S, Lees-Miller SP & Bochkarev A Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J 21, 1855–1863 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bochkareva E, Belegu V, Korolev S & Bochkarev A Structure of the major single-stranded DNA-binding domain of replication protein A suggests a dynamic mechanism for DNA binding. EMBO J 20, 612–618 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shastrula PK, Rice CT, Wang Z, Lieberman PM & Skordalakes E Structural and functional analysis of an OB-fold in human Ctc1 implicated in telomere maintenance and bone marrow syndromes. Nucleic Acids Res 46, 972–984 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bryan C, Rice C, Harkisheimer M, Schultz DC & Skordalakes E Structure of the human telomeric Stn1–Ten1 capping complex. PLoS ONE 8, e66756 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lim C et al. The structure of human CST reveals a decameric assembly bound to telomeric DNA. Science 368, 1081–1085 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study uses cryo-EM to solve the structure of the CST complex and reveals that ssDNA binding can trigger its assembly into a 2-MDa decameric supercomplex.
- 133.Bhattacharjee A, Wang Y, Diao J & Price CM Dynamic DNA binding, junction recognition and G4 melting activity underlie the telomeric and genome-wide roles of human CST. Nucleic Acids Res 45, 12311–12324 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hom RA & Wuttke DS Human CST prefers G-rich but not necessarily telomeric sequences. Biochemistry 56, 4210–4218 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen LY, Majerska J & Lingner J Molecular basis of telomere syndrome caused by CTC1 mutations. Genes Dev 27, 2099–2108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gu P et al. CTC1–STN1 coordinates G- and C-strand synthesis to regulate telomere length. Aging Cell 17, e12783 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Feng X et al. CTC1–STN1 terminates telomerase while STN1–TEN1 enables C-strand synthesis during telomere replication in colon cancer cells. Nat. Commun 9, 2827 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fan J & Pavletich NP Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev 26, 2337–2347 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gao Y et al. Structures and operating principles of the replisome. Science 363, eaav7003 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ge Y et al. Structural insights into telomere protection and homeostasis regulation by yeast CST complex. Nat. Struct. Mol. Biol 27, 752–762 (2020). [DOI] [PubMed] [Google Scholar]
- 141.Wan B et al. The Tetrahymena telomerase p75–p45–p19 subcomplex is a unique CST complex. Nat. Struct. Mol. Biol 22, 1023–1026 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lue NF et al. The telomere capping complex CST has an unusual stoichiometry, makes multipartite interaction with G-tails, and unfolds higher-order G-tail structures. PLoS Genet 9, e1003145 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gelinas AD et al. Telomere capping proteins are structurally related to RPA with an additional telomere-specific domain. Proc. Natl Acad. Sci. USA 106, 19298–19303 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sun J et al. Structural bases of dimerization of yeast telomere protein Cdc13 and its interaction with the catalytic subunit of DNA polymerase α. Cell Res 21, 258–274 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rice C & Skordalakes E Structure and function of the telomeric CST complex. Comput. Struct. Biotechnol. J 14, 161–167 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zeng Z et al. Structural basis for Tetrahymena telomerase processivity factor Teb1 binding to single-stranded telomeric-repeat DNA. Proc. Natl Acad. Sci. USA 108, 20357–20361 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Goulian M, Heard CJ & Grimm SL Purification and properties of an accessory protein for DNA polymerase α/primase. J. Biol. Chem 265, 13221–13230 (1990). [PubMed] [Google Scholar]
- 148.Zhao Y et al. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell 138, 463–475 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kratz K & de Lange T Protection of telomeres 1 proteins POT1a and POT1b can repress ATR signaling by RPA exclusion, but binding to CST limits ATR repression by POT1b. J. Biol. Chem 293, 14384–14392 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wan M, Qin J, Songyang Z & Liu D OB fold-containing protein 1 (OBFC1), a human homolog of yeast Stn1, associates with TPP1 and is implicated in telomere length regulation. J. Biol. Chem 284, 26725–26731 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tesmer VM, Smith EM, Danciu O, Padmanaban S & Nandakumar J Combining conservation and species-specific differences to determine how human telomerase binds telomeres. Proc. Natl Acad. Sci. USA 116, 26505–26515 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wu P, Takai H & de Lange T Telomeric 3′ overhangs derive from resection by Exo1 and Apollo and fill-in by POT1b-associated CST. Cell 150, 39–52 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Feng X, Hsu SJ, Kasbek C, Chaiken M & Price CM CTC1-mediated C-strand fill-in is an essential step in telomere length maintenance. Nucleic Acids Res 45, 4281–4293 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Diotti R, Kalan S, Matveyenko A & Loayza D DNA-directed polymerase subunits play a vital role in human telomeric overhang processing. Mol. Cancer Res 13, 402–410 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Porcella SY et al. Separable, Ctf4-mediated recruitment of DNA polymerase α for initiation of DNA synthesis at replication origins and lagging-strand priming during replication elongation. PLoS Genet 16, e1008755 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kilkenny ML et al. The human CTF4-orthologue AND-1 interacts with DNA polymerase α/primase via its unique C-terminal HMG box. Open. Biol 7, 170217 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nakaoka H, Nishiyama A, Saito M & Ishikawa F Xenopus laevis Ctc1–Stn1–Ten1 (xCST) protein complex is involved in priming DNA synthesis on single-stranded DNA template in Xenopus egg extract. J. Biol. Chem 287, 619–627 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ganduri S & Lue NF STN1–POLA2 interaction provides a basis for primase-pol α stimulation by human STN1. Nucleic Acids Res 45, 9455–9466 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Nunez-Ramirez R et al. Flexible tethering of primase and DNA pol α in the eukaryotic primosome. Nucleic Acids Res 39, 8187–8199 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Leteurtre F et al. Accelerated telomere shortening and telomerase activation in Fanconi’s anaemia. Br. J. Haematol 105, 883–893 (1999). [DOI] [PubMed] [Google Scholar]
- 161.Ball SE et al. Progressive telomere shortening in aplastic anemia. Blood 91, 3582–3592 (1998). [PubMed] [Google Scholar]
- 162.Shay JW & Wright WE Telomeres and telomerase: three decades of progress. Nat. Rev. Genet 20, 299–309 (2019). [DOI] [PubMed] [Google Scholar]
- 163.Savage SA Beginning at the ends: telomeres and human disease. F1000Res 10.12688/f1000research.14068.1 (2018). [DOI] [PMC free article] [PubMed]
- 164.Podlevsky JD, Bley CJ, Omana RV, Qi X & Chen JJ The telomerase database. Nucleic Acids Res 36, D339–D343 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Guo Y et al. Inherited bone marrow failure associated with germline mutation of ACD, the gene encoding telomere protein TPP1. Blood 124, 2767–2774 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bisht K, Smith EM, Tesmer VM & Nandakumar J Structural and functional consequences of a disease mutation in the telomere protein TPP1. Proc. Natl Acad. Sci. USA 113, 13021–13026 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Keller RB et al. CTC1 mutations in a patient with dyskeratosis congenita. Pediatr. Blood Cancer 59, 311–314 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Anderson BH et al. Mutations in CTC1, encoding conserved telomere maintenance component 1, cause Coats plus. Nat. Genet 44, 338–342 (2012). [DOI] [PubMed] [Google Scholar]
- 169.Simon AJ et al. Mutations in STN1 cause Coats plus syndrome and are associated with genomic and telomere defects. J. Exp. Med 213, 1429–1440 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Gu P & Chang S Functional characterization of human CTC1 mutations reveals novel mechanisms responsible for the pathogenesis of the telomere disease Coats plus. Aging Cell 12, 1100–1109 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wang Y & Chai W Pathogenic CTC1 mutations cause global genome instabilities under replication stress. Nucleic Acids Res 46, 3981–3992 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Maciejowski J & de Lange T Telomeres in cancer: tumour suppression and genome instability. Nat. Rev. Mol. Cell Biol 18, 175–186 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Makarov VL, Lejnine S, Bedoyan J & Langmore JP Nucleosomal organization of telomere-specific chromatin in rat. Cell 73, 775–787 (1993). [DOI] [PubMed] [Google Scholar]
- 174.Tommerup H, Dousmanis A & de Lange T Unusual chromatin in human telomeres. Mol. Cell. Biol 14, 5777–5785 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wu P & de Lange T No overt nucleosome eviction at deprotected telomeres. Mol. Cell. Biol 28, 5724–5735 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Galati A et al. The human telomeric protein TRF1 specifically recognizes nucleosomal binding sites and alters nucleosome structure. J. Mol. Biol 360, 377–385 (2006). [DOI] [PubMed] [Google Scholar]
- 177.Galati A et al. TRF2 controls telomeric nucleosome organization in a cell cycle phase-dependent manner. PLoS ONE 7, e34386 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Soman A et al. The human telomeric nucleosome displays distinct structural and dynamic properties. Nucleic Acids Res 48, 5383–5396 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ichikawa Y, Morohashi N, Nishimura Y, Kurumizaka H & Shimizu M Telomeric repeats act as nucleosome-disfavouring sequences in vivo. Nucleic Acids Res 42, 1541–1552 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.James TC et al. Distribution patterns of Hp1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur. J. Cell Biol 50, 170–180 (1989). [PubMed] [Google Scholar]
- 181.Gottschling DE, Aparicio OM, Billington BL & Zakian VA Position effect at S. cerevisiae telomeres—reversible repression of Pol II transcription. Cell 63, 751–762 (1990). [DOI] [PubMed] [Google Scholar]
- 182.Garcia-Cao M, O’Sullivan R, Peters AH, Jenuwein T & Blasco MA Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat. Genet 36, 94–99 (2004). [DOI] [PubMed] [Google Scholar]
- 183.Arnoult N, Van Beneden A & Decottignies A Telomere length regulates TERRA levels through increased trimethylation of telomeric H3K9 and HP1α. Nat. Struct. Mol. Biol 19, 948–956 (2012). [DOI] [PubMed] [Google Scholar]
- 184.Blasco MA The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet 8, 299–309 (2007). [DOI] [PubMed] [Google Scholar]
- 185.Dejardin J & Kingston RE Purification of proteins associated with specific genomic loci. Cell 136, 175–186 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Grolimund L et al. A quantitative telomeric chromatin isolation protocol identifies different telomeric states. Nat. Commun 4, 2848 (2013). [DOI] [PubMed] [Google Scholar]
- 187.Pfeiffer V, Crittin J, Grolimund L & Lingner J The THO complex component Thp2 counteracts telomeric R-loops and telomere shortening. EMBO J 32, 2861–2871 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ribes-Zamora A, Indiviglio SM, Mihalek I, Williams CL & Bertuch AA TRF2 interaction with Ku heterotetramerization interface gives insight into c-NHEJ prevention at human telomeres. Cell Rep 5, 194–206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Fisher TS & Zakian VA Ku: a multifunctional protein involved in telomere maintenance. DNA Repair 4, 1215–1226 (2005). [DOI] [PubMed] [Google Scholar]
- 190.Celli GB, Denchi EL & de Lange T Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat. Cell Biol 8, 885–U162 (2006). [DOI] [PubMed] [Google Scholar]
- 191.McKay SJ & Cooke H hnRNP A2/B1 binds specifically to single stranded vertebrate telomeric repeat TTAGGGn. Nucleic Acids Res 20, 6461–6464 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Ishikawa F, Matunis MJ, Dreyfuss G & Cech TR Nuclear proteins that bind the pre-mRNA 3′ splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Mol. Cell Biol 13, 4301–4310 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.LaBranche H et al. Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat. Genet 19, 199–202 (1998). [DOI] [PubMed] [Google Scholar]
- 194.Potjer TP et al. Multigene panel sequencing of established and candidate melanoma susceptibility genes in a large cohort of Dutch non-CDKN2A/CDK4 melanoma families. Int. J. Cancer 144, 2453–2464 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Pelusi S et al. Rare pathogenic variants predispose to hepatocellular carcinoma in nonalcoholic fatty liver disease. Sci. Rep 9, 3682 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Guacci A et al. Identification of a novel truncating mutation in PALB2 gene by a multigene sequencing panel for mutational screening of breast cancer risk-associated and related genes. J. Clin. Laboratory Anal 32, e22418 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Aoude LG et al. Nonsense mutations in the shelterin complex genes ACD and TERF2IP in familial melanoma. J. Natl Cancer Inst 107, dju408 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Vulliamy T et al. Telomere length measurement can distinguish pathogenic from non-pathogenic variants in the shelterin component, TIN2. Clin. Genet 81, 76–81 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sasa GS, Ribes-Zamora A, Nelson ND & Bertuch AA Three novel truncating TINF2 mutations causing severe dyskeratosis congenita in early childhood. Clin. Genet 81, 470–478 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Vulliamy TJ et al. Differences in disease severity but similar telomere lengths in genetic subgroups of patients with telomerase and shelterin mutations. PLoS ONE 6, e24383 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Robles-Espinoza CD et al. POT1 loss-of-function variants predispose to familial melanoma. Nat. Genet 46, 478–481 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Polvi A et al. Mutations in CTC1, encoding the CTS telomere maintenance complex component 1, cause cerebroretinal microangiopathy with calcifications and cysts. Am. J. Hum. Genet 90, 540–549 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Netravathi M et al. Whole exome sequencing in an Indian family links Coats plus syndrome and dextrocardia with a homozygous novel CTC1 and a rare HES7 variation. BMC Med. Genet 16, 5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Walne AJ et al. Mutations in the telomere capping complex in bone marrow failure and related syndromes. Haematologica 98, 334–338 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]



