Abstract
Reactive oxygen and nitrogen species (RONS) may infringe on the passing of pristine genetic information by inducing DNA inter- and intra-strand crosslinks, protein-DNA crosslinks, and chemical alterations to the sugar or base moieties of DNA. 8-Oxo-7,8-dihydroguanine (8-oxoG) is one of the most prevalent DNA lesions formed by RONS and is repaired through the base excision repair (BER) pathway involving the DNA repair glycosylases OGG1 and MUTYH in eukaryotes. MUTYH removes adenine (A) from 8-oxoG:A mispairs, thus mitigating the potential of G:C to T:A transversion mutations from occurring in the genome. The paramount role of MUTYH in guarding the genome is well established in the etiology of a colorectal cancer predisposition syndrome involving variants of MUTYH, referred to as MUTYH-associated polyposis. In this review, we highlight recent advances in understanding how MUTYH structure and related function participate in the manifestation of human disease such as MAP. Here we focus on the importance of MUTYH’s metal cofactor sites, including a recently discovered “Zinc linchpin” motif, as well as updates to the catalytic mechanism. Finally, we touch on the insight gleaned from studies with MAP-associated MUTYH variants and recent advances in understanding the multifaceted roles of MUTYH in the cell, both in the prevention of mutagenesis and tumorigenesis.
Keywords: Base excision repair, glycosylase, 8-oxoguanine, MutY, MUTYH, MUTYH-associated polyposis, Fe-S clusters
Graphical abstract
1. Introduction: MutY’s Nobel Heritage
In the late 1980s, the laboratory of 2015 Chemistry Nobel Laureate Paul Modrich discovered an activity in E. coli that was able to restore G:A mismatches to G:C base-pairs [1]. This activity was independent of methylation status of the template strand, and therefore could not be attributed to mismatch repair (MMR). Around the same time, Miller and co-workers identified a mutator locus in E. coli that generates G:C to T:A transversion mutations, which they termed as the mutY gene [2]. Together Miller and Modrich determined that the mutY gene product was an adenine glycosylase [3], similar to the base excision repair (BER) Uracil-DNA glycosylase (UDG) [4], discovered by 2015 Chemistry Nobel Laureate Tomas Lindahl [5]. Using a combination of biochemistry and genetics, Miller, Michaels and co-workers demonstrated that MutY plays an important role in the prevention of mutations caused by the oxidatively damaged guanine lesion, 8-oxo-7,8-dihydroguanine (8-oxoG) by removing adenine (A) from 8-oxoG:A base pairs that form due to the miscoding properties of 8-oxoG. [2, 6-10]. The knowledge of these fundamental features of MutY were crucial in the discovery of a colorectal cancer (CRC) predisposition syndrome involving variants of the human MutY homolog, MUTYH, in a disease referred to as MUTYH-associated polyposis (MAP) [11, 12].
The importance of MUTYH in MAP, and increased interest in targeting DNA repair enzymes as new therapeutic strategies, have upped the ante in understanding the inherent chemistry of MutY/MUTYH and BER glycosylases [13]. In this review, we will highlight recent insights into the structural and functional properties of MutY, MUTYH and MAP variants. For example, recent investigations of MAP variants in the David laboratory resulted in the discovery of a second metal binding site in mammalian homologs of MUTYH [14]. Furthermore, despite MutY’s discovery almost 30 years ago, new features of MutY base excision have been revealed prompting revisions to the accepted mechanism for MutY, and implicating that similar revisions may be appropriate for related glycosylases [15]. We also look beyond MAP and call attention to MUTYH’s roles in other diseases, including intriguing links between MUTYH and neurological disorders [16-19]. These various aspects of MUTYH collectively accentuate how the structure and function of MutY and MUTYH elegantly tie to their activities in the cell and the ever-unraveling roles they play in protecting life from DNA damage. Of note, in this review, we highlight primarily new studies and therefore direct the reader to previous reviews on MutY, MUTYH and MAP for additional details [12, 20-27].
2. Oxidatively Damaged DNA and Base Excision Repair
2.1 Oxidatively damaged DNA and 8-oxoG
Replicating cells continue life by passing on genetic information in the form of DNA. If the information within DNA is compromised by DNA base modifications resulting from reactions with chemical species, such as reactive oxygen and nitrogen species (RONS), malignant cellular phenotypes may result, ultimately leading to diseases such as cancer [28-31]. RONS arise from cellular exposure to xenobiotic toxins, as well as ionizing radiation and ultraviolet light. However, RONS are more frequently formed as a consequence of cellular respiration, inflammatory responses, and by-products of cellular signaling and lipid peroxidation [reviewed by 32].
DNA damage resulting from RONS manifests in the form of DNA inter- and intra-strand crosslinks, protein-DNA crosslinks, and chemical alterations to the sugar or base moieties [12, 33, 34]. The most well studied oxidative base modification to DNA is 8-oxoG, which occurs within the human genome approximately once per million guanine residues [12, 35, 36]. The prevalence of 8-oxoG in genomic DNA is due to the low redox potential of guanine, making it highly susceptible to oxidation [37-39]. The correlation between oxidative stress and high levels of 8-oxoG has resulted in the use of 8-oxoG as a cellular biomarker [40-42].
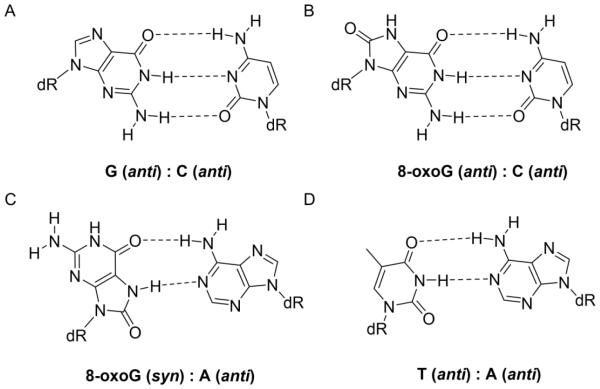
The formation of 8-oxoG via oxidation results in the addition of an oxo group at the C8 position and a hydrogen atom at the N7 position of guanine (Figure 1). When 8-oxoG is oriented in the normal anti conformation within DNA, the 8-oxo group sterically clashes and unfavorably interacts with the O4’ position of the DNA sugar. Because of this, 8-oxoG preferentially adopts the syn conformation during DNA replication offering a Hoogsteen face that resembles that of the Watson-Crick face of thymine [43, 44]. Because of this dual-coding ability, replicative polymerases often misincorporate A opposite 8-oxoG [12, 45]. If this 8-oxoG:A mispair persists within DNA without being repaired, a G:C to T:A transversion mutation permanently incorporates into the genome. For example, G:C to T:A mutations have been shown to be the predominant type of somatic mutation in protein kinase genes in lung, breast and colorectal cancers [46].
Figure 1.
The DNA contexts of 8-oxoG. When a normal G:C base pair (A) is damaged to form an 8-oxoG:C base pair (B), 8-oxoG may adopt a syn conformation about its glycosidic bond to form a stable 8-oxoG:A base pair after a round of replication (C). Following a subsequent round of replication, where A is in the template DNA strand, a T:A base pair results (D).
2.2 The GO repair pathway
Tomas Lindahl’s insight that the high level of spontaneous cytosine deamination to uracil must be repaired in order to preserve life resulted in the seminal discovery of UDG and the BER pathway [5, 47-49]. The BER pathway mitigates the mutagenic and toxic properties of a wide array of DNA nucleobase modifications [12, 50]. The key players of this pathway are a cadre of DNA glycosylases that scour the genome and initiate the repair of specific types of base modifications in DNA by hydrolyzing the N-glycosidic bond between the damaged base and the 2’-deoxyribose sugar, affording apurinic/apyrimidinic (AP) sites within DNA [4]. Mono-functional glycosylases pass the AP site to a 5’ AP endonuclease (such as APE1 in humans) that hydrolyzes the phosphodiester DNA backbone at the AP site. Further trimming of the remaining sugar fragments allows for gap-filling by a DNA polymerase to replace the excised nucleotide. Lastly, DNA ligase provides the finishing touches of repair by sealing the phosphodiester backbone [51]. The mammalian BER pathway is more complex than its bacterial counterpart and has two distinct sub pathways: short-patch and long-patch BER, which differ in the number of nucleotides incorporated during the polymerase step and the various enzymes involved [52-55].
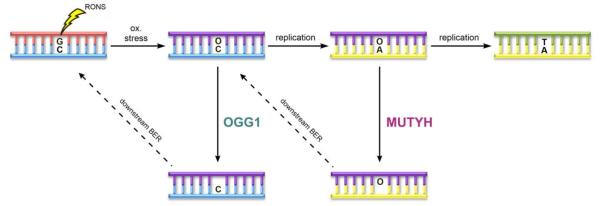
Miller and Michaels coined the term “GO” (for “Guanine Oxidation”) repair pathway to describe the combined efforts of MutM (Fpg), MutY and MutT in mitigating the mutagenic properties of 8-oxoG in bacteria (Figure 2) [7, 8, 10, 12, 56]. Later work established a similar GO repair pathway in human cells featuring the functional equivalents OGG1, MUTYH and NUDT1 [57-62]. The first responder to the scene to cope with 8-oxoG in the human genome is the 8-oxoG Glycosylase (OGG1), which is a bifunctional glycosylase that recognizes 8-oxoG:C base pairs, cleaves out 8-oxoG, and catalyzes subsequent β-elimination to afford a nick in the DNA backbone 5’ to the nascent AP site [63]. If 8-oxoG:C base pairs escape OGG1 excision and 8-oxoG-containing DNA is used as a template for DNA replication, the miscoding nature of 8-oxoG leads to formation of 8-oxoG:A base pairs. Notably, this damaged and mismatched base pair is stable within the DNA helix, and often eludes the DNA proofreading activities of DNA polymerases [25, 45, 64]. MUTYH plays a crucial role at this stage by excising the misplaced A before another round of replication ensues and seals the G:C to T:A mutation within the genome. Following the action of MUTYH, subsequent processing by downstream BER machinery recreates an 8-oxoG:C base pair, providing another opportunity for OGG1 to excise substrate 8-oxoG lesions [12].
Figure 2.
The “GO” repair pathway. Depiction of the GO repair pathway, featuring the key base excision responsibilities of OGG1 and MUTYH.
The last player in the GO Repair pathway, Nudix hydrolase or NUDT1 (formally known as MutT human homolog 1, or MTH1), is recruited by the cell to hydrolyze 8-oxoG nucleoside triphosphates to monophosphates to prevent damaged nucleotide incorporation into the genome [65]. If NUDT1 fails to sanitize the 8-oxoGTP pool, 8-oxoG:A lesions may form where A is in the contextually correct template DNA strand. If A were cleaved in this context, the action of MUTYH would be pro-mutagenic, highlighting the importance of NUDT1 within the cell.
3. The MutY Mechanism in Depth
DNA repair glycosylases are responsible for recognizing specific types of DNA damage and performing subsequent base excision catalysis, and must function appropriately to complete repair and maintain genomic integrity [4]. The necessary role of MUTYH in preventing mutations and associated diseases has been well established, and much information has been provided on the details of how MutY enzymes recognize 8-oxoG:A mismatches and catalyze base excision [12]. However, new insights into the activity of MutY enzymes continue to be discovered. Detailing the chemistry of MutY’s “search and rescue” mission will help researchers gain understanding of the etiology of MAP, as well as determine the relative cellular impacts of different variants. Detailed knowledge of the mechanism of enzymes similar to DNA glycosylases has previously led to development of small molecule inhibitors, paving the way for the design of new pharmaceutical drugs [66]. Furthermore, the idea of targeting DNA repair enzymes as a therapeutic strategy is gaining ground within the cancer research community [13, 67, 68].
3.1 Structural features of MutY enzymes and 8-oxoG:A recognition
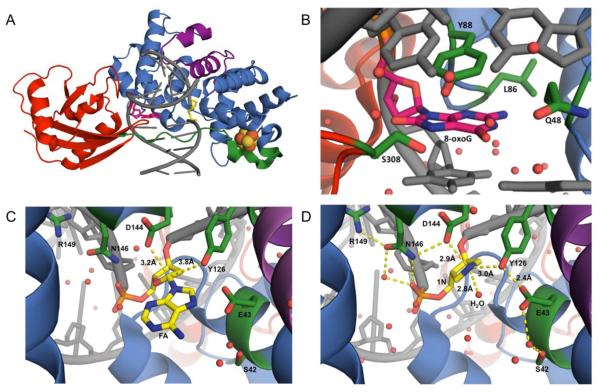
MutY enzymes are members of the structurally related BER superfamily of glycosylases that contain a signature helix-hairpin-helix DNA binding motif followed by a glycine-proline rich region and a conserved catalytic aspartic acid residue (HhH-GPD motif) [4]. The N-terminal domain harbors an adenine-specific binding pocket, the catalytic residues required for base excision, and the [4Fe-4S]2+ cluster (vide infra) [4, 9, 12, 69]. In addition, MutY enzymes differ from other BER glycosylases in harboring a unique C-terminal domain that plays an important role in 8-oxoG recognition and exhibits structural homology to NUDT1 [4, 12, 70-76]. The N- and C-terminal domains are united by a flexible interdomain connector (IDC). Crystal structures of a catalytically inactive D144N Geobacillus stearothermophilus (Gs) MutY bound to an 8-oxoG:A-containing duplex (Lesion recognition complex or LRC) [77], WT Gs MutY bound to a cleavage resistant substrate analogue, arabino 2'-fluoro-2'-deoxyadenosine (FA) (Fluorine Lesion Recognition Complex or FLRC) [78], and WT Gs MutY bound to an azaribose transition state (TS) analog (1N) (Transition State Analog Complex or TSAC) [15] (Figure 3) provide insight into the structural features used in damage DNA recognition and base excision. Key features present in all three structures include Gs MutY remodeling of the DNA that induces a kink at the lesion site and a DNA bend of approximately 50 degrees. The HhH and [4Fe-4S]2+ cluster loop (FCL) DNA binding motifs within the catalytic N-terminal domain make extensive contacts with the FA/A/1N-containing strand. The FA/A/1N nucleotide is rotated out of the helix, and in the structures with FA/A, the base is positioned in the active site pocket. In addition, both the C- and N-terminal domains make extensive contacts with the 8-oxoG-containing strand. Of note, the 8-oxoG nucleotide remains in the DNA helical stack, with an anti N-glycosidic bond conformation.
Figure 3.
Structural insight into the mechanism of MutY. (A) Crystal structure of prokaryotic Gs MutY (PDB ID: 5DPK) bound to DNA (grey) containing 8-oxoG (hot pink) opposite the azasugar transition state mimic, 1N (yellow). The N-terminal domain of MutY (blue), contains the catalytic machinery for adenine excision, including the signature HhH-GPD motif (purple) and the [4Fe-4S]2+ cluster (orange and yellow spheres). The NUDT1-like C-terminal domain of MutY (red) primarily serves in 8-oxoG recognition. The IDC (green) of bacterial homologs of MutY wraps across the DNA major groove. (B) Close up of 8-oxoG (hot pink) in the active site of MutY. 8-oxoG experiences extensive interactions with MutY, stabilizing the damaged base (interacting amino acids are labeled and colored in forest green). Comparison of the substrate base analogs (C) FA (of PDB ID: 3G0Q, FLRC crystal structure) and (D) 1N (of PDB ID: 5DPK, TSAC crystal structure) within the active site of Gs MutY. Amino acid residues of MutY that are in close proximity to the FA/1N (yellow) nucleotides are labeled and colored in forest green. The position of the potential catalytic water molecule within close proximity to N1’ of 1N is labeled as H2O in panel D. In panels B, C, D, water molecules are depicted as red spheres, distances are demonstrated by yellow dashes, and elements are depicted with oxygen in red, nitrogen in blue, and phosphorous in orange.
3.2 Features influencing the location of 8-oxoG:A mismatches by MutY enzymes
Details regarding the early steps in the process by which MutY enzymes locate rare 8-oxoG:A mismatches, while effectively rejecting structurally similar T:A base pairs, remain somewhat murky. MutY, like many other glycosylases, uses a one-dimensional search process, likely combining a series of sliding and hopping motions to search the genome for 8-oxoG:A mismatches [79, 80]. It is evident that key early steps in MutY damage recognition involve contacts with 8-oxoG, in order to “select” for an inappropriately positioned A. Indeed, a truncated form of E. coli (Ec) MutY lacking the C-terminal domain exhibits reduced glycosylase activity, and lacks enhanced affinity for 8-oxoG containing duplexes compared to the WT enzyme [74, 81]. Moreover, the absence of the C-terminal domain with MutY diminishes the repair of an 8-oxoG:A plasmid substrate within a cellular context [82]. Notably, despite the ability of MutY enzymes to catalyze adenine removal from G:A and 8-oxoG:A substrates in vitro, minimal cellular repair on G:A-containing plasmid substrates was observed, again pointing to the critical nature of 8-oxoG recognition for selecting appropriate contexts for adenine excision [71]. These cellular assays demonstrate that base pair discrimination is more stringent in vivo, consistent with the additional demands of locating rare mismatches amongst a vast excess of undamaged DNA.
Structural studies of Fpg interrogating normal DNA revealed the importance of a Phe “wedge” residue that intercalates into the DNA helix, thus aiding in the location of 8-oxoG:C base pairs [12, 83-85]. In addition, single-molecule visualization of quantum-dot labeled F111A Fpg showed an absence of slow-moving enzymes and a faster enzyme diffusion rate compared to the WT enzyme [86]. These observations are consistent with the idea that this large aromatic residue plays an important role in the damage interrogation process. Notably, a similar role may be ascribed to a highly conserved Tyr residue in MutY enzymes. In the LRC/FLRC/TSAC structures, Tyr 88 (Tyr 165 in human MUTYH) is wedged between 8-oxoG and its 5’-neighboring base, disrupting base-stacking interactions that stabilize the lesion in duplex DNA (Figure 3B) [77, 78]. The intercalation of the Tyr wedge likely promotes the “flipping” of its substrate adenine into the enzyme’s active site. In addition, the Tyr wedged into the DNA stack stabilizes the bent DNA, and thereby decreases the activation barrier to catalysis [77, 87]. The importance of Tyr 165 is highlighted in MAP, where inherited mutations at this position are of the most common leading to disease pathogenesis [12, 88].
Locating 8-oxoG is only half of the task of MutY enzymes since discrimination between 8-oxoG:A and 8-oxoG:C mispairs is important to avoid wasting time and inappropriately acting upon the “wrong” substrate. In a recent structure of Gs MutY interrogating an 8-oxoG:C base pair, the non-target cytosine base is flipped out of the helix into an “exo” site for initial recognition; this exo-site is structurally separate from the adenine-processing active site [89]. Using point mutations, Verdine and co-workers showed that this exo-site can be structurally disrupted allowing the extrahelical cytosine to be flipped into the active site. However the anti-substrate cytosine is not cleaved due to repulsive interactions caused by the enzyme’s catalytic aspartate residue. Indeed, the final verification checkpoint is the active site that is tailored to catalyze adenine base removal [90]. Together, the combined features of initial recognition of 8-oxoG, and proper engagement of only adenine in the active site afford the enzyme with a line of defense against non-specific repair during its normal search and interrogation process.
3.3 New insights into the MutY mechanism
Functionally, MutY is similar to UDG in that both enzymes are monofunctional glycosylases that catalyze hydrolysis of an N-glycosidic bond to afford AP sites within DNA [4]. For several years there existed a heated debate surrounding whether or not MutY acted as a bifunctional glycosylase with the ability to catalyze an associated β-lyase strand scission reaction at the AP site product [4]. However, the rate of adenine base excision was found to far outpace the rate of strand scission; moreover, the extent of strand scission provided by the enzyme was considerably less than that provided by base quenching [91]. The observed weak β-lyase activity of MutY [92, 93] appears to be a result of opportunistic reactions with nearby lysine residues found within the active site due in part to the high affinity of MutY for the AP site product [4, 94]. Indeed, in Ec MutY, mutation of the Lys residue (Lys 142) responsible for making the transient Schiff base intermediate with the AP site did not diminish the glycosylase activity [95-97].
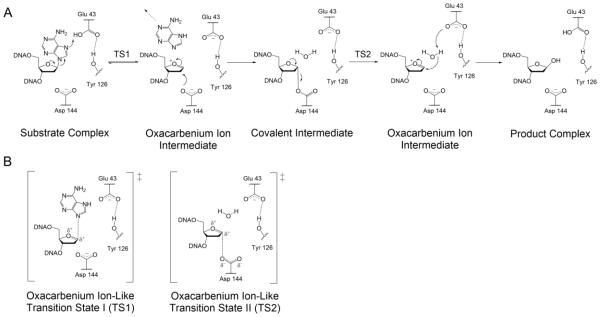
Kinetic isotope effect (KIE) studies on MutY with a G:A substrate indicated that, like UDG, MutY performs catalysis through a stepwise SN1 (DN*AN‡) mechanism involving an oxacarbenium ion intermediate [78, 98]. The FLRC structure of Gs MutY (Figure 3C), along with structural, kinetic and mutagenesis studies, suggest that a Glu residue (E43 in Gs MutY, E120 in MUTYH) participates in catalysis as a general acid, while an Asp residue (D144 in Gs MutY, D222 in MUTYH) stabilizes the build-up of positive charge in the oxacarbenium ion intermediate [69, 77, 78, 82, 99]. The recently published structure of Gs MutY bound to an azaribose transition state analog (TSAC) provides a definitive picture of the key active site players necessary for glycosidic bond hydrolysis [15]. In the TSAC structure, the Tyr 126 residue is within hydrogen-bonding distance to the positively charged N1’ of 1N implicating a role for this residue in catalysis. Tyr 126 was also implicated to participate in MutY catalysis in computational studies [100]. Moreover, the single amino acid variant Y126F was found to have dramatically reduced A excision activity compared to WT [15]. Notably, Tyr 126 (Ser 120 in Ec MutY) is at the same amino acid position as the catalytic Lys residues found in bifunctional BER glycosylases (such as OGG1); in fact, S120K Ec MutY is a bifunctional glycosylase/lyase, albeit with a reduced rate for the glycosylase activity [101].
A surprising feature of the TSAC structure is the presence of a water molecule in the space occupied by the adenine base in the FLRC structure, and in close proximity to the N1’ of 1N in the TSAC structure (Figure 3). It was anticipated that a potential nucleophilic water would be located near Asp 144, on the other side of the sugar nucleotide, poised for nucleophilic attack at the oxacarbenium ion intermediate [15]. This suggested that if the water molecule in the TSAC structure represents the nucleophilic water, the base must depart completely before the water gains access to the active site. The ability of MutY to utilize methanol in its adenine excision chemistry provided the opportunity to test this mechanistic proposal based on the TSAC structure using two-dimensional NMR experiments. The MutY-methanolysis product was necessary for these experiments because stereochemical information of the water-mediated reaction is lost due to the equilibration of α- and β-anomers of the abasic site hemiacetal in solution. Through this work, our laboratory established that only the β-anomer of the methanolysis adduct is formed by Gs MutY establishing retention of stereochemistry in the MutY mechanism.
These results led our laboratory to propose a revised mechanism for MutY comprising two nucleophilic displacement steps similar to the accepted mechanism of “retaining” O-glycosidases (Figure 4) [102-105]. Once the 8-oxoG:A base pair has been located, and the adenine base has been inserted into the MutY active site, protonation of N7 by Glu 43 facilitates cleavage of the N-glycosidic bond to form an oxacarbenium ion intermediate in an SN1-like fashion consistent with KIE studies [98, 106]. Asp 144 likely plays an important role in stabilizing the positive charge build-up during the glycosidic bond cleavage, and then rapidly reacts with the oxacarbenium ion to form the transient acetal covalent intermediate. After formation of the acetal intermediate, adenine vacates the active site, allowing room for a nucleophilic water molecule to diffuse into the active site. Presumably, the second hydrolysis step also proceeds via an SN1-like mechanism to generate a second highly reactive transient oxacarbenium ion intermediate. The active site Glu 43 is positioned to activate the water molecule for nucleophilic attack at the C1’ carbon of the acetal intermediate forming the β-anomer of the final abasic site product [15]. Both the Glu and Asp residues have been found to be essential for catalysis, and when these residues are mutated to remove the carboxylate (such as to Gln or Asn, respectively) the enzyme activity is completely abolished [69, 77, 82, 99]. Interestingly when the Glu residue was replaced with an Asp, the activity was severely diminished, yet when the Asp was converted to a larger Glu residue, the enzyme retained WT-like activity. The reduced activity of E43D Ec MutY is likely due to the shorter side chain being less efficient at protonation of N7 and activating the water nucleophile [82]. In addition, when the catalytic Asp was mutated to a Cys residue, reduced activity was observed with a shift in the pH profile consistent with the thiolate mimicking the role of the Asp carboxylate. These results suggest the importance of a residue at the position of the catalytic Asp (Cys or Glu) capable of forming a covalent bond with the oxacarbenium ion intermediate.
Figure 4.
Revised mechanism for MutY-catalyzed adenine excision. (A) The recently proposed double displacement mechanism for MutY enzymes based on methanolysis studies that revealed retention of configuration at the anomeric carbon. This result suggests the involvement of a covalent intermediate, most likely involving Asp 144, in analogy to similar work with retaining O-glycosidases. (B) The oxacarbenium ion-like transition states formed during the proposed MutY mechanism. For both TSs, a highly dissociative transition state based on previous KIE measurements is shown; the extent of participation of the nucleophile in the two TSs remains to be established.
Following catalysis, MutY stays tightly bound to its abasic site product, and dissociation of the enzyme-product DNA complex is extremely slow [73, 107, 108]. In conjunction with a lack of AP lyase activity, MutY enzymes have evolved to “hold on” to the abasic site DNA product and protect it from potential strand scission before being handed off to an AP Endonuclease to continue the repair process [4]. Turnover of Ec MutY is enhanced by the addition of the bacterial AP endonuclease Endo IV [109]. Similarly, APE1 has been shown to increase turnover, and stimulate mammalian MutY 8-oxoG:A activity, and this interaction is mediated by direct protein-protein contacts [108, 110, 111]. Collectively the synergistic action of both MUTYH and APE1 ensures that faithful repair is performed without producing single-strand DNA breaks (SSB’s), which under accumulation may signal cell death.
4. MUTYH-Associated Polyposis
4.1 Discovery of MAP and clinical features
In 2002, a groundbreaking study reported a unique case of three siblings within a British family (Family N) that showed clinical symptoms of familial adenomatous polyposis (FAP), an inherited predisposition mechanism of CRC characterized by the formation of polyps lining the epithelial tissue of the large intestine [11]. FAP is caused by constitutional heterozygous mutations in the APC gene, the gene product of which is the Adenomatous Polyposis Coli (APC) protein, a cellular regulator of colon cell proliferation [112]. Due to the known familial association of the polyposis phenotype and inherited mutations in APC characteristic of FAP, it was surprising when DNA sequencing of Family N failed to uncover any inherited pathogenic APC mutations. Notably, however, sequencing of the APC gene from the tumors of the affected family members revealed a high proportion of G:C to T:A transversion mutations; this same type of mutation accumulates in MutY- and Ogg1-deficient strains of bacteria [2, 8, 113, 114]. Suspecting faulty action within the GO repair pathway, sequencing of the MUTYH, OGG1 and NUDT1 genes from lymphoblastoid cells uncovered that the affected members of Family N possessed inherited biallelic germline mutations in the MUTYH gene, encoding missense variants Y165C and G382D MUTYH.
In vitro analysis of the corresponding E. coli variants Y82C and G253D MutY showed that both variants were catalytically compromised when compared to the intrinsic activity of wild-type MutY [11]. Reduced activity was observed for G:A substrates, with the rate of adenine excision at 37 °C reduced by roughly 98% and 86% for Y82C and G253D, respectively. In addition, reactions with 8-oxoG:A lesions measured at 2 °C also revealed the catalytic defects of both variants; the glycosylase activity of G253D was decreased by 85% when compared to WT, whereas the activity of Y82C was comprised to such an extent that minimal substrate processing was observed. These functional assays supported the hypothesis that the polyposis phenotype present in Family N was a consequence of inefficient repair by the inherited MUTYH variants of 8-oxoG:A bps in the APC gene, leading to its inactivation. Since then, many efforts have demonstrated the causal link between the inheritance of MUTYH variants and colorectal polyposis [115-120] leading to classification of a distinct mechanism to early onset CRC, now referred to as MUTYH-associated polyposis [121] [118].
The MAP phenotype most closely resembles that of attenuated FAP (AFAP), both of which are less severe compared to FAP in terms of the number of colorectal polyps, age of onset and age of developing CRC [11, 115, 122]. Currently, patients are diagnosed with MAP at a mean age of 45 years and harbor 15-100 colorectal polyps. MAP patients within 50 and 80 years of age have a 20-80% chance of developing CRC. Generally, age of onset in MAP is later than either FAP or Lynch Syndrome (LS), an autosomal dominant CRC syndrome caused by inherited DNA MMR deficiencies [123-125]. The highly similar phenotypes of FAP, MAP and LS can be difficult to distinguish clinically. Indeed, some 10-15% of suspected LS cases, characterized by tumors with microsatellite instability and lack of MMR protein expression, cannot be explained by mutation analysis of MMR genes alone [126, 127]. For example, a patient displaying clinical symptoms of LS was found to harbor somatic MSH2 mutations leading to microsatellite instability and resulting CRC. Upon closer inspection, the patient was found to possess biallelic germline mutations in the MUTYH gene that led to the accumulation of somatic transversion mutations in MSH2, KRAS and APC. These were the first reported findings of MAP mimicking LS, further highlighting the role of malfunctioning MUTYH in cancer [128].
4.2 From genetics to structure and function: Y165C and G382D
After the discovery of MAP, further analysis of Y82C and G253D Ec MutY revealed that both variants have diminished binding affinities for 8-oxoG:FA mispairs in vitro. Moreover, the mutant MutY enzymes lack the ability to distinguish between G and 8-oxoG in duplex DNA [129]. A structural basis for the compromised enzymatic activity of Y165C was provided by the Gs MutY LRC/FLRC structures, where the corresponding Tyr (Tyr 88 in Gs MutY, Tyr 82 in Ec MutY) intercalates 5’ of the aberrant 8-oxoG lesion, and hydrogen bonds with the N7 hydrogen of 8-oxoG [77, 78]. The intercalation or “wedging” role is likely important for locating the 8-oxoG:A mismatch based on work with the corresponding Phe residue in Fpg (vide supra). Furthermore, this intercalation role appears to be more important for stabilizing the necessary catalytic conformation, rather than simply making the hydrogen bond to NH7, as was demonstrated via temperature-dependent glycosylase assays of a series of mutated Y82X Ec MutY enzymes (where X = Phe, Cys, or Leu); these studies revealed the ability of Tyr to be effectively replaced by Leu without loss of activity, while substitution with a smaller Cys residue resulted in substantial increases in the activation barrier to catalysis [87]. Gly 253 is localized in a tight turn region of the protein that hydrogen bonds via the peptide amide backbone to phosphodiester of the 8-oxoG nucleotide [77, 78]. Replacement of the Gly residue with a larger residue such as Asp or Ala likely destabilizes the tight turn and reduces the extent of engagement with the 8-oxoG:A base pair [87]. These structural and functional studies illustrate the critical roles played by both residues in recognizing 8-oxoG:A base pairs, a prerequisite for adenine base excision.
Detailed kinetic analysis of the adenine glycosylase activity of MUTYH variants Y165C and G328D has been difficult due to the low levels of overexpression and quality of the human protein. Indeed, this has resulted in considerable discrepancies between previous reports on the activity of the two human variants [130-132]. These discrepancies are likely due, in part, to the absence of correction for differences in active fraction, as the active fraction can vary considerably even among different preparations of the same WT enzyme [131, 133]. Expression of a maltose-binding protein fusion of WT, Y165C and G382D MUTYH in insect cells provides higher quality protein for detailed kinetic and post translational modification analysis [88, 133]. Using this approach, G382D MUTYH expressed at levels similar to the WT enzyme, while Y165C routinely expressed with active fractions that were 10% of the WT enzyme [88]. Rate constants for adenine excision measured under single-turnover conditions (correcting for active fraction) indicated an activity for G382D that is modestly reduced (60% of WT) while Y165C exhibited minimal activity. Of note, the glycosylase activities of the human variants in this work matched well with the previous studies on the corresponding variants in the bacterial and murine homologs [87, 88, 111, 129, 131]. Despite the distinct differences in the in vitro kinetics, Mutyh-null mouse embryonic fibroblasts (MEFs) expressing Y165C and G382D MUTYH revealed that both forms mediate similarly low levels of repair of an 8-oxoG:A base pair in a green fluorescent protein (GFP) reporter plasmid [88]. These experiments illustrate that the activity of G382D MUTYH is more dramatically reduced within cells than the in vitro experiments indicated. This may be due in part to the more difficult task of repair in a cellular context [82, 87, 134], as well as other features that may come into play in a cellular context, such as interactions and competition with other proteins [88, 111]. These studies underscore the importance of multiple assays, along with structural studies, to fully explore the functional implications of specific variants.
4.3 Q324H: MAP variant or harmless polymorph?
The clinical side of MAP has moved rapidly; over 300 MUTYH variants have been reported in the Leiden Open Variant Database (LOVD), many of which are missense variants with no data on functional consequences [135]. A recent study of a large number of DNA samples from colorectal cancer patients found that one in 45 individuals are MUTYH mutation carriers [136]; these estimates were similar to those from previous studies [137]. Notably, only 12 well-characterized MUTYH variants (such as Y165C, G382D, and several truncating mutations) were tested for, and this prompts concern that such studies may underestimate the prevalence of MUTYH mutations. Another complicating feature is that some MUTYH variants are thought to be harmless polymorphisms and therefore an individual may be incorrectly categorized as “no risk”, even if they are a monoallelic carrier. This underscores the need for fully understanding the functional consequences of different variants in order to properly diagnose patients and assess potential CRC risk.
The complications associated with designation of polymorph versus cancer variant is particularly well-illustrated with Q324H MUTYH, a variant that has long been considered a common polymorphism, and is particularly high in prevalence in some ethnic populations [138]. However, there have also been genetic studies that suggest an increased risk for CRC and lung cancer associated with the Q324H variant [139-142]. Analysis of the adenine glycosylase activity of Q324H MUTYH indicated a slightly reduced activity, similar to that observed with G382D MUTYH. Using the GFP-based cellular repair assay described above, our lab showed that 8-oxoG:A repair mediated by Q324H MUTYH was close to that of Y165C and G382D [88]. Using a similar approach, Mazzei and co-workers have found that expression of Q324H in cells was associated with 2-fold higher levels of 8-oxoG in DNA, hypersensitivity to oxidants and accumulation of the population in the S phase of the cell cycle [143]. Q324H is found in the IDC of MUTYH, an area of the protein not directly involved in DNA binding or catalysis, but known to facilitate key repair protein interactions with the Rad9-Hus1-Rad1 (9-1-1) complex and APE1 [134]. Q324H was shown to have reduced affinity for Hus-1 of the 9-1-1 complex compared to the WT protein [134, 144]. Importantly, altered interactions with protein partners such as the 9-1-1 complex may result in MAP due to improper signaling responses to DNA damage [23, 143].
4.4 MAP variants with a myriad of consequences
The genetic, structural and functional data for Y165C and G382D MUTYH are all consistent with defective repair of 8-oxoG:A mismatches leading to CRC. With many other variants, as illustrated above with Q324H, the data is not quite as compelling. Indeed, in some cases, a variant may be exceedingly rare such that the correlation with CRC cannot be convincingly made on the basis of genetics. The location of some MAP variants may predict dysfunction, however, the location and type of many missense variants are not obviously detrimental to function. Not all variants have been tested by a battery of assays, and by several laboratories, as has been the case with Y165C and G382D MUTYH. A recent review article catalogued the glycosylase activity of variants that have been reported [27]. There are many variants that have been reported to have WT or moderately reduced glycosylase activity, similar to the results with G382D MUTYH. In several cases, assays using the ability of the human MUTYH variant to complement for the lack of Ec MutY in suppressing spontaneous mutations have been used to gauge activity [129, 131, 145]. For example, Kundu et al. showed that the MAP variants P391L and Q324R had similar defects in glycosylase activity in vitro [131], however, their ability to prevent mutations in MutY- cells were starkly different; P391L was found to exhibit negligible ability to suppress mutations while the Q324R variant s0075ppressed mutations like the WT enzyme [131]. This again underscores the different functional properties of different variants, and the need to perform multiple types of analyses to gauge function.
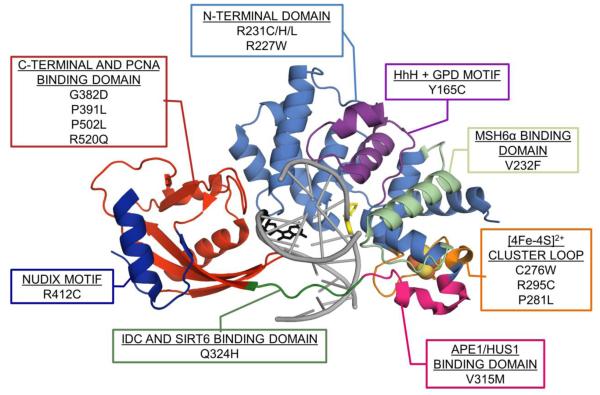
Of note, MAP-associated variants have also helped to reveal the importance of domains outside those involved in 8-oxoG recognition and adenine base excision (Figure 5). Indeed, the Q324H/R variants helped to illustrate the importance of the IDC in mediating protein-protein interactions that play an important role in DNA repair and the DNA damage response [14, 88, 143]. Similarly, the MAP-associated variant V315M has been shown to weaken a critical 9-1-1 complex binding interaction to MUTYH [146]. Modified amino acids in the PCNA-binding region of MUTYH, such as P502L and R520Q, have been reported in MAP patients [107, 147, 148]. Functional studies in vitro have shown that P502L has similar DNA binding and glycosylase activity to WT enzyme, while binding affinity interactions with PCNA are dramatically reduced, which may more dramatically affect cellular repair. R520Q on the other hand was shown to have significantly reduced substrate binding, glycosylase activity and PCNA binding relative to WT [134].
Figure 5.
MAP map. Crystal structure of prokaryotic Gs MutY (PDB ID: 5DPK) bound to DNA (light grey) containing 8-oxoG (black) opposite 1N (yellow). MAP variants and domains discussed in this review are indicated on the MAP map and correspond to the MUTYH homologous positioning on the structure of MutY. *Note, for consistency with previous work, the amino acid positions are based on the 535 amino acid MUTYH isoform α-3 (UniProt ID# Q9UIF7-3). The positions of MUTYH MAP variants in the LOVD are categorized both by the longer 549 MUTYH isoform numbering (such that Y165C corresponds to Y179C), as well as the numbering of the shorter aforementioned isoform.
5. MUTYH and its Metal Cofactors
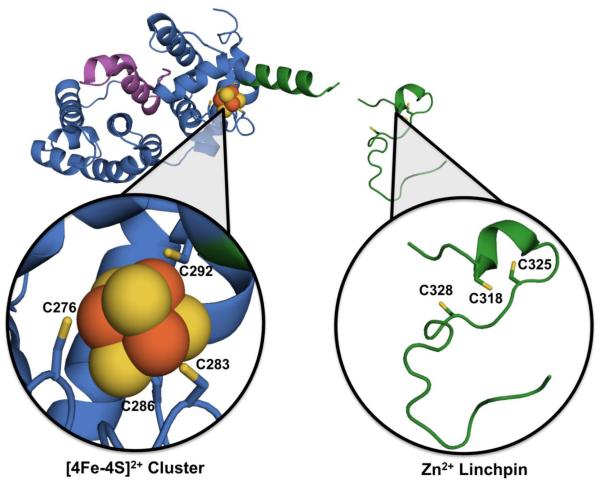
MUTYH harbors two vital cofactors, both of which are located remotely from the active site (Figure 6). One of these cofactors, a [4Fe-4S]2+ (Fe-S) cluster, is chelated by four Cys residues in the N-terminal domain, and positions an Fe-S cluster loop (FCL) motif that mediates interactions with the DNA substrate [69]. All but a few recently identified homolog lineages of MutY possess this iron-sulfur cluster [149]. The second cofactor is a recently identified Zn2+ ion, coordinated to three Cys residues in the interdomain connector (IDC) of higher eukaryotic MutY homologs; this region of MUTYH has been dubbed the “Zn2+ linchpin motif” to embody its structural and functional roles [14]. These two MUTYH cofactors are essential for efficient repair in cells, but their exact role in DNA repair and regulation is still under investigation [14, 134, 150, 151].
Figure 6.
The fragment crystal structure of H. sapien MUTYH (PDB ID: 3N5N) lacking the C-terminal 8-oxoG recognition domain, with close-ups to metal binding sites. Located in the N-terminal catalytic domain of MUTYH (blue) is the [4Fe-4S]2+ cluster (Fe2+/3+ in orange spheres and sulfide in yellow spheres, with Cys ligands as sticks, sulfur highlighted also in yellow) as well as the HhH GPD motif (purple). Linking the N- and C-terminal domains is the IDC (green), which contains the Zn2+ linchpin motif (Cys ligands responsible for coordinating Zn2+ metal shown as sticks, sulfur highlighted in yellow) and is conserved amongst higher eukaryotic homologs.
5.1 The essential role(s) of the Fe-S cluster within MUTYH
Iron-Sulfur clusters are structurally and functionally diverse cofactors, found in a variety of DNA binding enzymes including helicases, polymerases, and glycosylases [152, 153]. The presence of the Fe-S cluster in the bacterial homolog MutY was originally anticipated due to its high sequence similarity to a bacterial glycosylase that acts on damaged pyrimidine bases known as Endonuclease III (EndoIII) [9]. Like EndoIII, the Fe-S cluster of MutY is coordinated to four highly conserved cysteinyl ligands with a spacing of Cys-X6-Cys-X2-Cys-X5-Cys [9, 152]. Refolding experiments of MutY either in the presence or absence of Fe(II) and S2− demonstrated that cluster assembly is not needed for MutY re-folding; however, importantly, the refolded forms lacking the cluster were unable to bind to substrate DNA, but the affinity could be rescued by Fe-S reconstitution following refolding [154]. These studies underscore the importance of the proper coordination of the region around the Fe-S cluster and the FCL motif in order to mediate key electrostatic interactions between positive amino acids of the FCL and the negative DNA backbone [69, 150]. Replacement of the Ec MutY cluster coordinating Cys ligands using Site Directed Mutagenesis (SDM) with His, Ser or Ala showed higher levels of expression for a fully–coordinated Fe-S cluster and revealed that one of the Cys ligands anchoring the FCL motif could be replaced with a His ligand (C199H Ec MutY). Notably, C199H behaved similarly to WT in terms of glycosylase activity and mutation suppression but displayed an enhanced affinity for substrate DNA further implicating this region in substrate DNA binding [151, 155].
Fe-S clusters are ideally suited to participate in electron transfer processes [156], however MutY and MUTYH would not be expected to need redox chemistry for glycosidic bond cleavage based on the known mechanism of MutY (vide supra). Electrochemical assays utilizing DNA-modified gold electrodes demonstrated that the Fe-S cluster of Ec MutY is able to redox cycle with a midpoint potential of +90 mV versus NHE, similar to that observed for High-potential (HiPIP) Fe-S cluster proteins [157]. Consistent with the signal arising from the Fe-S cluster, C199H MutY exhibited a signal with a shifted potential [157]. This study also established that the electrochemical signal is sensitive to perturbations in the DNA helix, such as the presence of an AP site [157]. The importance of DNA binding for the redox chemistry and the similarity in redox potential of Fe-S clusters in several BER glycosylases led the Barton laboratory to propose a novel hypothesis that electron exchange between repair enzymes via DNA-mediated charge transport (CT) may serve as a mechanism to locate DNA damage [157-161]. This damage location strategy relies on the integrity of the DNA helix to facilitate the CT between enzymes distally separated. DNA damage and mispairs attenuate DNA-mediated CT, causing the DNA repair enzyme to remain engaged on DNA and continue its genome-wide search for damage [162, 163].
5.2 MAP variants surrounding the Fe-S cluster
Thirteen known MAP-associated MUTYH variants are near the Fe-S cluster and the FCL signifying the impact of this region on MUTYH repair: R295C, P281L, C276W, R227W/Q, V232F, R233X, R231C/G/H/L, and G272E/X (Figure 5). While some of these variants have been studied in detail, other variants are hardly mentioned in the literature [27, 119, 134, 164]. Some studies have generated contradictory results for the same variant, which causes the pathogenicity of certain variants to be disputed [165]. Of the variants listed, R295C has been heavily studied, and demonstrated to have reduced affinity for damaged DNA, slower rate of adenine excision and significant reduction in active fraction compared to wild type [134]. Furthermore, R295C retained Hus1 affinity despite the variant’s location within the 9-1-1 complex binding domain [134]. This result contradicted a previous study [166], leading to the consideration that some variants’ defects may be more subtle than others which, as a result, could require a more detailed investigation of adenine glycosylase kinetics to reveal the true extent of any defects that are present [134]. Indeed, the results of the functional assays with R295C MUTYH are reminiscent of those described above with G382D MUTYH, where the defects in the glycosylase assay are subtle, and dependent on the conditions of the assay [88].
The variant P281L was shown to have extremely compromised substrate DNA binding affinity, resulting in minimal levels of adenine excision and very low levels of active MUTYH [134]. The Pro residue lies within the solvent exposed and highly conserved FCL motif of MUTYH, and the low level of activity is consistent with the key role the FCL motif plays in damaged substrate recognition. The tendency of Pro residues to introduce kinks, in conjunction with the bulkiness of a Leu residue, suggests that the Leu replacement results in significant conformational changes to the FCL that effectively restrict its ability of effectively engage an 8-oxoG:A mismatch. The R227W MUTYH variant is also quite interesting in that the corresponding Arg in the bacterial counterparts was implicated to be close to DNA on the basis of photo-crosslinking [167] and the LRC structure [77]. Notably, Arg 227 is also located within the hMSH6 binding domain of MUTYH. This variant displays severe defects in 8-oxoG:A processing yet has no effect on the physical interaction with hMSH6 [168]. V232F also shows defects in 8-oxoG:A lesion binding and glycosylase activity yet this variant’s interaction with hMSH6 is unchanged. Using a bacterial complementation assay, both human variants exhibited a reduced ability to prevent DNA mutations relative to WT MUTYH [168].
R233X and C276W variants have yet to be evaluated for their effects on MUTYH-mediated repair. Only brief mentions of the discovery of these two variants exist in the literature [169, 170]. R233X is a nonsense mutation first discovered in a compound heterozygote polyp patient that also had the Y165C mutation in the MUTYH gene [170]. Given that R233X is a nonsense mutation, the likely protein product is a truncated protein that lacks the Fe-S cluster. Cys 276 corresponds to one of the Cys ligands of the Fe-S cluster that is part of the FCL. Replacement of this Cys with a Trp would be expected to alter activity, potentially by destabilizing the Fe-S cluster. Of note, previous SDM on the Cys ligands in Ec MutY (vide infra) showed a large difference depending on which Cys residue was substituted and the nature of the substitution. Moreover, the results indicated that replacement of the Cys ligands of the FCL was more readily tolerated than the other Cys ligands [151].
Several of the cluster-associated variants have population-dependent frequencies, such as R231C/H/L and G272E [171-173]. R231H is present in Swiss populations and both R231C and G272E are present in Japanese populations. Notably, the R231H/L and G272E variants were demonstrated to have severely reduced DNA binding and repair activity [171-173]. Curiously, the R231C variant may provide a new potential ligand that could alter the native cluster coordination, impacting enzyme functionality. Alternatively, Arg 231 may be another positively charged-residue in this region that is participating in interactions with the DNA phosphodiester backbone, given the variants spatial proximity to the FCL.
5.3 Discovery of a novel Zn2+ linchpin within the IDC of MUTYH
Connecting the C- and N-terminal domains of MUTYH is the fifty-six amino acid long IDC. Interestingly, the IDC of MUTYH is significantly longer than that of its bacterial homologs [14]. Furthermore, H. sapien MUTYH shares 78% sequence homology to M. musculus Mutyh and only 41% sequence homology to Ec MutY [61, 62, 174]. Previous studies have revealed that this evolved region within eukaryotic MUTYH homologs participates in interactions with DNA repair response enzymes [107, 108, 175-177]. The presence of the “Zinc Linchpin” motif of the IDC was initially suggested by the observation of three highly conserved Cys residues within the IDC of MUTYH mammalian homologs with a spacing of Cys-X6-Cys-X2-Cys [14]. Metal analysis of M. Musculus Mutyh revealed the presence of a single Zn2+ ion in addition to the four Fe atoms of the N-terminal Fe-S cluster [14]. EXAFS analysis of M. musculus Mutyh revealed that the Zn2+ ion is most likely tetra-coordinated either to four sulfur atoms, or three sulfur and one-two oxygen/nitrogen-containing ligands [14]. Surprisingly, however, in the crystal structure of a truncated form of MUTYH (Figure 6) the three conserved Cys are in a disordered region of the protein, and there is an absence of a coordinated Zn2+ ion [177]. It is likely that the Zn2+ ion was lost during purification or crystallization. Importantly, SDM of the Zn2+ coordinating Cys ligands to Ser or Ala significantly reduced the active fraction of enzyme in vitro, suggesting that the Zn2+ linchpin plays an important role in engaging the DNA substrate. In addition, the Cys ligand mutations that resulted in loss of the coordinated Zn2+ ion also resulted in the loss of the ability to complement for the lack of MutY in suppressing mutations in bacteria. Notably, replacement of the two key Cys residues required for Zn2+ coordination with Ser (C325S and C328S MUTYH) greatly reduced the Fe-S cluster retention as demonstrated by UV/vis absorbance measurements and metal analysis [14]. This suggests that the two metal cofactor sites are not only important for MUTYH function, but that their functions may be intimately connected to each other.
5.4 MAP variants within the IDC
The Zn2+ ion is coordinated to Cys residues located near several known protein docking sites within the IDC of MUTYH [14]. One of these protein partners is the receiver of the MUTYH AP site product, APE1 [107, 110]. In addition, the Zn2+ linchpin is located within the 9-1-1 complex binding domain [146, 177]. Both MUTYH and the 9-1-1 complex participate in an ATR-mediated Chk1 activation cell signal cascade [178] [179]. The mutated Gln of the MAP variant Q324H, is also within the Zn2+ binding site, and this variant was shown to have reduced interaction with the 9-1-1 complex as discussed previously [134]. The importance of Zn2+ coordination for MUTYH repair is evident, however further studies evaluating the importance of the metal ion for interactions with protein partners and potential consequences due to adjacent MAP variants are needed. One of the Zn2+ linchpin ligands has also been recorded as a MAP variant (C318R); the potential consequence of changing the metal chelating ligand to that of a basic residue warrants further experimentation [180]. Remarkably, histone deacetylase protein SIRT6 has also been shown to interact with MUTYH in a region of the IDC that also contains MAP variants, and this interaction also deserves further inquiry [181]. Many more of the IDC MAP variants need a quantitative functional assessment as their contribution to pathogenicity still remains unknown.
6. Beyond MAP: Other implications of MUTYH in Medicine
6.1 A Balancing Act: MUTYH prevents cancer through suppressing mutations or inducing cell death
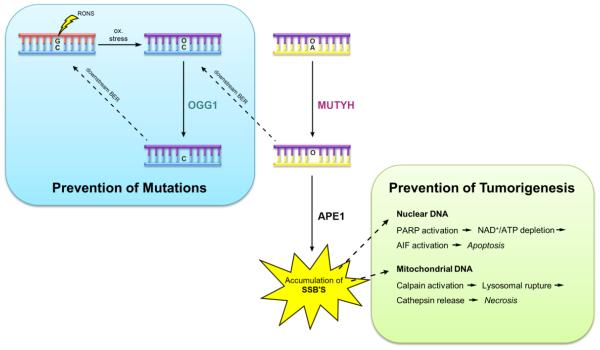
Following the actions of NUDT1 and OGG1, MUTYH is the third line of defense against the deleterious effects of 8-oxoG. However, it takes the action of all three enzymes to prevent accumulation of 8-oxoG within DNA. It has previously been shown that Ogg1-deficient and Mth1-deficient mice accumulate 8-oxoG in mitochondrial and nuclear DNA [182, 183]. As 8-oxoG:A lesions build up after replication, Mutyh expression was shown to cause cell death [23, 184, 185]. Indeed, Ogg1/Mth1 double-KO mice do not develop lung tumors, whereas Mutyh-KO mice have the most significant increase in spontaneous tumor formation when compared to Ogg1-KO and Mth1-KO mice [186, 187]. When OGG1 is preferentially expressed in either the mitochondria or nucleus of Ogg1-KO MEFs, 8-oxoG can selectively accumulate in mitochondrial or nuclear DNA. As 8-oxoG:A lesions form in either DNA, Mutyh-mediated adenine excision generates a massive build up of AP sites, which can then form SSB’s after the action of an AP endonuclease [184]. In the nucleus, poly (ADP-ribose) polymerase (PARP) activation acts as a molecular SSB sensor, depleting cellular ATP and NAD+ concentrations, as well as activating apoptosis inducing factor (AIF), both of which lead to cell death. In mitochondria, the buildup of SSB’s activates calpains which induce lysosomal rupture ultimately leading to cell death (Figure 7). siRNA knockdown of Mutyh has been shown to efficiently suppress SSB formation in nuclear and mitochondrial DNA [184].
Figure 7.
Schematic illustrating the roles of MUTYH in preventing carcinogenesis. MUTYH activity prevents of mutations associated with 8-oxoG by removing A from 8-oxoG:A mismatches (blue). However, under conditions that produce overwhelming numbers of 8-oxoG:A lesions, the A excision activity of MUTYH, and subsequent product of strand breaks provides signals that lead to cell death that prevent tumorigenesis (green).
Similar findings have also been reported in bacteria, where Foti et al. demonstrated that the toxicity of beta-lactams and quinolones, oxidative bactericidal antibiotics, is due to the incomplete repair of double-strand breaks formed from closely spaced 8-oxoG lesions. Lethality of the antibiotics were suppressed in cells lacking MutT (NUDT1 homolog) and MutY [188].
By implementing MUTYH, the cell has evolved to avoid the pass-down of mutated genetic formation and subsequent tumorigenesis in two ways: by helping to restore G:C base-pairs that may otherwise be lost due to 8-oxoG formation, and by helping the cell execute cell death in the event that the genome is beyond faithful repair. Indeed, it has recently been reported that p53 acting as a transcription factor targets the MUTYH gene, thus regulating MUTYH expression as a mediator of p53 tumor suppression [189]. Collectively, these findings establish clear roles for MUTYH in the prevention of cancer. However, recent evidence has shown that MUTYH overexpression may be detrimental and contribute to the pathogenesis of neurological disorders.
6.2 The Dark Side: MUTYH and neurodegenerative diseases
Increased oxidative stress and 8-oxoG formation are hallmark characteristics of neurodegenerative disorders such as Alzheimer’s, Huntington’s and Parkinson’s disease [17-19]. Not surprisingly, altered gene expression profiles for glycosylases involved in the GO Repair pathway have been shown to be linked to disease pathogenesis [16-19]. Expression levels of NUDT1, OGG1 and MUTYH, for instance, have been shown to be upregulated in the mitochondria of dopaminergic neurons in the brains of Parkinson’s disease patients [190].
In 2012, Sheng et al. showed that OGG1- and NUDT1-deficient microglia and neurons selectively accumulate high levels of 8-oxoG in mitochondrial DNA and nuclear DNA, respectively [191]. In these cells, MUTYH expression led to an accumulation of SSBs and subsequent activation of PARP in nuclear DNA, and calpains in mitochondrial DNA. Both mechanisms trigger cell death ultimately contributing to neurodegeneration. MUTYH-induced neurodegeneration was pronounced such that Ogg1/Mutyh double-KO cells exhibited significantly higher resistance to oxidation-induced striatal degeneration when compared to Ogg1 KO and WT cells [191].
7. Concluding Remarks and Perspectives
The 2015 Nobel Prize in Chemistry was awarded to Tomas Lindahl, Paul Modrich and Aziz Sancar for “having mapped, at the molecular level, how cells repair damaged DNA and safeguard the genetic information” [192]. Indeed, the work of Lindahl and Modrich led to the discovery of MutY, and its unusual activity as an adenine glycosylase. These discoveries paved the way for uncovering the important role of MutY and MUTYH in preventing mutations associated with 8-oxoG, and the discovery of MAP. The role of MUTYH variants in MAP underscores the harmful nature of oxidatively damaged DNA, and the importance of mechanisms for maintaining the genome in the face of continued oxidative insults. Despite the large amount of work on delineating the structural and functional properties of MutY, new features have recently been unveiled. One of these new discoveries was uncovering the presence of a previously unrecognized Zn2+ ion in the IDC of MUTYH, and demonstrating a critical role for the Zn2+ linchpin motif in damage recognition and repair. This discovery, along with the many new studies of Fe-S clusters in BER glycosylases, and other DNA repair enzymes, prompt additional work on determining how these metal sites may be utilized during MUTYH-mediated DNA damage response, and the potential impact of MAP variants on metal cofactor function. In addition, the discovery that MutY is a retaining glycosidase prompts additional studies to further define this mechanism, and to determine if similar mechanisms are operative in other BER glycosylases. Moreover, these mechanistic features may be exploited to design new therapeutic strategies to target MUTYH. Lastly, the details by which MutY finds 8-oxoG:A mismatches is still unclear, and the exact defects of many MAP variants are also a mystery. We envision that structural, biophysical and mechanistic studies of MUTYH and MAP variants will be even more needed going forward. Lastly, we anticipate that new discoveries of the role of MUTYH in human disease will come from many directions, such as those capitalizing on new advances in DNA sequencing, as well as probing gene functions in cells with new gene editing tools.
Highlights.
MutY is a unique BER glycosylase that removes adenine from 8-oxoG:A mispairs
Dysfunctional MUTYH variants are associated with MUTYH-associated polyposis (MAP)
There are many MAP variants that have a myriad of functional consequences
*MUTYH contains a recently identified Zinc-linchpin motif that is required for efficient mismatch recognition and repair
New structural and mechanistic information indicated that MutY is a retaining glycosidase
Acknowledgements
We are particularly grateful for the wonderful cadre of people that have passed through the David laboratory, and have helped in building our part of the MutY/MUTYH story piece by piece. We especially thank Drs. Jeremy Cheadle and Julian Sampson for recruiting us to work with them in the discovery of MAP. We also thank Dr. Martin Horvath for our on-going collaboration that led to the Gs MutY TSAC structure. Our work on the topics of this review was funded by the National Cancer Institute (CA067985). Doug Banda and Katie Bradshaw were supported by a National Institute of Environmental Health Sciences funded training program in Environmental Health Sciences (T32 ES007058). We apologize for not citing all of the work on MutY and MUTYH due to limitations on space and time.
Abbreviations
- G
guanine
- C
cytosine
- A
adenine
- T
thymine
- MMR
mismatch repair
- BER
base excision repair
- UDG
uracil DNA glycosylase
- 8-oxoG
8-oxo-7,8-dihydroguanine
- CRC
colorectal cancer
- MUTYH
MutY homolog
- MAP
MUTYH-associated polyposis
- RONS
reactive oxygen and nitrogen species
- AP
apurinic-apyrimidinic or abasic
- APE1
apurinic/apyrimidinic endonuclease
- Fpg
formamidopyrimidine DNA glycosylase
- OGG1
8-oxoguanine glycosylase 1
- NUDT1
nudix hydroylase
- IDC
interdomain connector
- Gs
Geobacillus Stearothermophilus
- Ec
Escherichia coli
- LRC
lesion recognition complex
- WT
wild type
- FA
arabino 2'-fluoro-2'-deoxyadenosine
- FLRC
fluorine lesion recognition complex
- HhH
helix-hairpin-helix
- FCL
iron-sulfur cluster loop
- TSAC
transition state analog complex
- 1N
azaribose transition state analog
- TS
transition state
- KIE
kinetic isotope effect
- SSB
single-strand break
- FAP
familial adenomatous polyposis
- APC
adenomatous polyposis coli
- LS
Lynch syndrome
- MSH2
MutS homolog 2
- MEF
mouse embryonic fibroblast
- GFP
green fluorescent protein
- PCNA
proliferating cell nuclear antigen
- RPA
replication protein A
- 9-1-1
Rad9-Hus1-Rad1
- SDM
site directed mutagenesis
- CT
charge transport
- hMSH6
human MutS homolog 6
- PARP
poly (ADP-ribose) polymerase
- AIF
apoptosis inducing factor
- KO
knock out
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Su SS, Lahue RS, Au KG, Modrich P. Mispair specificity of methyl-directed DNA mismatch correction in vitro. The Journal of biological chemistry. 1988;263(14):6829–35. [PubMed] [Google Scholar]
- [2].Nghiem Y, Cabrera M, Cupples CG, Miller JH. The mutY gene: a mutator locus in Escherichia coli that generates G.C----T.A transversions. Proceedings of the National Academy of Sciences of the United States of America. 1988;85(8):2709–13. doi: 10.1073/pnas.85.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Au KG, Clark S, Miller JH, Modrich P. Escherichia coli mutY gene encodes an adenine glycosylase active on G-A mispairs. Proceedings of the National Academy of Sciences. 1989;86(22):8877–8881. doi: 10.1073/pnas.86.22.8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].David SS, Wiliams SD. Chemistry of glycosylases and endonucleases involved in base-excision repair. Chemical reviews. 1998;98(3):1221–1261. doi: 10.1021/cr980321h. [DOI] [PubMed] [Google Scholar]
- [5].Lindahl T. Uracil-DNA glycosylase from Escherichia coli. Methods Enzymol. 1980;65(1):284–90. doi: 10.1016/s0076-6879(80)65038-1. [DOI] [PubMed] [Google Scholar]
- [6].Au KG, Cabrera M, Miller JH, Modrich P. Escherichia coli mutY gene product is required for specific A-G----C.G mismatch correction. Proceedings of the National Academy of Sciences of the United States of America. 1988;85(23):9163–6. doi: 10.1073/pnas.85.23.9163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Michaels ML, Cruz C, Grollman AP, Miller JH. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proceedings of the National Academy of Sciences. 1992;89(15):7022–7025. doi: 10.1073/pnas.89.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Michaels ML, Miller JH. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) J Bacteriol. 1992;174(20):6321–5. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Michaels ML, Pham L, Nghiem Y, Cruz C, Miller JH. MutY, an adenine glycosylase active on G-A mispairs, has homology to endonuclease III. Nucleic Acids Research. 1990;18(13):3841–5. doi: 10.1093/nar/18.13.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Michaels ML, Tchou J, Grollman AP, Miller JH. A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry. 1992;31(45):10964–8. doi: 10.1021/bi00160a004. [DOI] [PubMed] [Google Scholar]
- [11].Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, Hodges AK, Davies DR, David SS, Sampson JR, Cheadle JR. Inherited variants of MYH associated with somatic G : C -> T : A mutations in colorectal tumors. Nature Genetics. 2002;30(2):227–232. doi: 10.1038/ng828. [DOI] [PubMed] [Google Scholar]
- [12].David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447(7147):941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pearl LH, Schierz AC, Ward SE, Al-Lazikani B, Pearl FMG. Therapeutic opportunities within the DNA damage response. Nat Rev Cancer. 2015;15(3):166–180. doi: 10.1038/nrc3891. [DOI] [PubMed] [Google Scholar]
- [14].Engstrom LM, Brinkmeyer MK, Ha Y, Raetz AG, Hedman B, Hodgson KO, Solomon EI, David SS. A zinc linchpin motif in the MUTYH glycosylase interdomain connector is required for efficient repair of DNA damage. J Am Chem Soc. 2014;136(22):7829–32. doi: 10.1021/ja502942d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Woods RD, O'Shea VL, Chu A, Cao S, Richards JL, Horvath MP, David SS. Structure and stereochemistry of the base excision repair glycosylase MutY reveal a mechanism similar to retaining glycosidases. Nucleic Acids Research. 2016;44(2):801–810. doi: 10.1093/nar/gkv1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Nakabeppu Y, Tsuchimoto D, Yamaguchi H, Sakumi K. Oxidative damage in nucleic acids and Parkinson's disease. J Neurosci Res. 2007;85(5):919–34. doi: 10.1002/jnr.21191. [DOI] [PubMed] [Google Scholar]
- [17].Polidori MC, Mecocci P, Browne SE, Senin U, Beal MF. Oxidative damage to mitochondrial DNA in Huntington's disease parietal cortex. Neuroscience letters. 1999;272(1):53–6. doi: 10.1016/s0304-3940(99)00578-9. [DOI] [PubMed] [Google Scholar]
- [18].Shimura-Miura H, Hattori N, Kang D, Miyako K, Nakabeppu Y, Mizuno Y. Increased 8-oxo-dGTPase in the mitochondria of substantia nigral neurons in Parkinson's disease. Annals of neurology. 1999;46(6):920–4. [PubMed] [Google Scholar]
- [19].Wang J, Xiong S, Xie C, Markesbery WR, Lovell MA. Increased oxidative damage in nuclear and mitochondrial DNA in Alzheimer's disease. Journal of neurochemistry. 2005;93(4):953–62. doi: 10.1111/j.1471-4159.2005.03053.x. [DOI] [PubMed] [Google Scholar]
- [20].de Oliveira AH, da Silva AE, de Oliveira IM, Henriques JA, Agnez-Lima LF. MutY-glycosylase: an overview on mutagenesis and activities beyond the GO system. Mutat Res. 2014;769:119–31. doi: 10.1016/j.mrfmmm.2014.08.002. [DOI] [PubMed] [Google Scholar]
- [21].Nielsen M, Lynch H, Infante E, Brand R, Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD, Fong CT, Mefford HC, Smith RJH, Stephens K. Gene Reviews. University of Washington, Seattle. All rights reserved; Seattle (WA): 2012. MUTYH-Associated Polyposis. [Google Scholar]
- [22].Sampson JR, Jones N. MUTYH-associated polyposis. Best Pract Res Clin Gastroenterol. 2009;23(2):209–18. doi: 10.1016/j.bpg.2009.03.006. [DOI] [PubMed] [Google Scholar]
- [23].Oka S, Nakabeppu Y. DNA glycosylase encoded by MUTYH functions as a molecular switch for programmed cell death under oxidative stress to suppress tumorigenesis. Cancer Sci. 2011;102(4):677–82. doi: 10.1111/j.1349-7006.2011.01869.x. [DOI] [PubMed] [Google Scholar]
- [24].Mazzei F, Viel A, Bignami M. Role of MUTYH in human cancer. Mutat Res. 2013;743-744:33–43. doi: 10.1016/j.mrfmmm.2013.03.003. [DOI] [PubMed] [Google Scholar]
- [25].Markkanen E, Dorn J, Hubscher U. MUTYH DNA glycosylase: the rationale for removing undamaged bases from the DNA. Front Genet. 2013;4:18. doi: 10.3389/fgene.2013.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].de Miranda NF, Hes FJ, van Wezel T, Morreau H. Role of the microenvironment in the tumourigenesis of microsatellite unstable and MUTYH-associated polyposis colorectal cancers. Mutagenesis. 2012;27(2):247–53. doi: 10.1093/mutage/ger077. [DOI] [PubMed] [Google Scholar]
- [27].Wallace SS, Murphy DL, Sweasy JB. Base excision repair and cancer. Cancer Lett. 2012;327(1-2):73–89. doi: 10.1016/j.canlet.2011.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kidane D, Chae WJ, Czochor J, Eckert KA, Glazer PM, Bothwell AL, Sweasy JB. Interplay between DNA repair and inflammation, and the link to cancer. Crit Rev Biochem Mol Biol. 2014;49(2):116–39. doi: 10.3109/10409238.2013.875514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].van Loon B, Markkanen E, Hübscher U. Oxygen as a friend and enemy: How to combat the mutational potential of 8-oxo-guanine. DNA repair. 2010;9(6):604–616. doi: 10.1016/j.dnarep.2010.03.004. [DOI] [PubMed] [Google Scholar]
- [30].Winterbourn CC. Reconciling the chemistry and biology of reactive oxygen species. Nat Chem Biol. 2008;4(5):278–86. doi: 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- [31].Kasai H, Crain PF, Kuchino Y, Nishimura S, Ootsuyama A, Tanooka H. Formation of 8-hydroxyguanine moiety in cellular DNA by agents producing oxygen radicals and evidence for its repair. Carcinogenesis. 1986;7(11):1849–51. doi: 10.1093/carcin/7.11.1849. [DOI] [PubMed] [Google Scholar]
- [32].West JD, Marnett LJ. Endogenous reactive intermediates as modulators of cell signaling and cell death. Chemical research in toxicology. 2006;19(2):173–94. doi: 10.1021/tx050321u. [DOI] [PubMed] [Google Scholar]
- [33].Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411(6835):366–74. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- [34].Wiseman H, Halliwell B. Damage to DNA by reactive oxygen and nitrogen species: role in inflammatory disease and progression to cancer. Biochemical Journal. 1996;313:17–29. doi: 10.1042/bj3130017. Pt 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Nakabeppu Y. Cellular levels of 8-oxoguanine in either DNA or the nucleotide pool play pivotal roles in carcinogenesis and survival of cancer cells. Int J Mol Sci. 2014;15(7):12543–57. doi: 10.3390/ijms150712543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Steenken S, Jovanovic SV. How Easily Oxidizable Is DNA? One-Electron Reduction Potentials of Adenosine and Guanosine Radicals in Aqueous Solution. Journal of the American Chemical Society. 1997;119(3):617–618. [Google Scholar]
- [37].Bruskov VI, Malakhova LV, Masalimov ZK, Chernikov AV. Heat-induced formation of reactive oxygen species and 8-oxoguanine, a biomarker of damage to DNA. Nucleic Acids Res. 2002;30(6):1354–63. doi: 10.1093/nar/30.6.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Neeley WL, Essigmann JM. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chemical research in toxicology. 2006;19(4):491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- [39].Ohno M, Miura T, Furuichi M, Tominaga Y, Tsuchimoto D, Sakumi K, Nakabeppu Y. A genome-wide distribution of 8-oxoguanine correlates with the preferred regions for recombination and single nucleotide polymorphism in the human genome. Genome Res. 2006;16(5):567–75. doi: 10.1101/gr.4769606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kasai H. Analysis of a form of oxidative DNA damage, 8-hydroxy-2'-deoxyguanosine, as a marker of cellular oxidative stress during carcinogenesis. Mutat Res. 1997;387(3):147–63. doi: 10.1016/s1383-5742(97)00035-5. [DOI] [PubMed] [Google Scholar]
- [41].Paz-Elizur T, Krupsky M, Elinger D, Schechtman E, Livneh Z. Repair of the oxidative DNA damage 8-oxoguanine as a biomarker for lung cancer risk. Cancer biomarkers : section A of Disease markers. 2005;1(2-3):201–5. doi: 10.3233/cbm-2005-12-308. [DOI] [PubMed] [Google Scholar]
- [42].Sliwinska A, Kwiatkowski D, Czarny P, Toma M, Wigner P, Drzewoski J, Fabianowska-Majewska K, Szemraj J, Maes M, Galecki P, Sliwinski T. The levels of 7,8-dihydrodeoxyguanosine (8-oxoG) and 8-oxoguanine DNA glycosylase 1 (OGG1) - A potential diagnostic biomarkers of Alzheimer's disease. Journal of the neurological sciences. 2016;368:155–9. doi: 10.1016/j.jns.2016.07.008. [DOI] [PubMed] [Google Scholar]
- [43].Culp SJ, Cho BP, Kadlubar FF, Evans FE. Structural and conformational analyses of 8-hydroxy-2'-deoxyguanosine. Chemical research in toxicology. 1989;2(6):416–22. doi: 10.1021/tx00012a010. [DOI] [PubMed] [Google Scholar]
- [44].Uesugi S, Ikehara M. Carbon-13 magnetic resonance spectra of 8-substituted purine nucleosides. Characteristic shifts for the syn conformation. Journal of the American Chemical Society. 1977;99(10):3250–3253. doi: 10.1021/ja00452a008. [DOI] [PubMed] [Google Scholar]
- [45].Beard WA, Batra VK, Wilson SH. DNA polymerase structure-based insight on the mutagenic properties of 8-oxoguanine. Mutat Res. 2010;703(1):18–23. doi: 10.1016/j.mrgentox.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O'Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Menzies A, Mironenko T, Perry J, Raine K, Richardson D, Shepherd R, Small A, Tofts C, Varian J, Webb T, West S, Widaa S, Yates A, Cahill DP, Louis DN, Goldstraw P, Nicholson AG, Brasseur F, Looijenga L, Weber BL, Chiew YE, DeFazio A, Greaves MF, Green AR, Campbell P, Birney E, Easton DF, Chenevix-Trench G, Tan MH, Khoo SK, Teh BT, Yuen ST, Leung SY, Wooster R, Futreal PA, Stratton MR. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446(7132):153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lindahl T. DNA glycosylases, endonucleases for apurinic/apyrimidinic sites, and base excision-repair. Prog Nucleic Acid Res Mol Biol. 1979;22:135–92. doi: 10.1016/s0079-6603(08)60800-4. [DOI] [PubMed] [Google Scholar]
- [48].Lindahl T. DNA repair enzymes. Annu Rev Biochem. 1982;51:61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- [49].Friedberg EC, Lindahl T. Inroads into base excision repair II. The discovery of DNA glycosylases. "An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues," Proc. Nat. Acad. Sci. USA, 1974. DNA repair. 2004;3(11):1532–6. doi: 10.1016/j.dnarep.2004.05.014. discussion 1531-2. [DOI] [PubMed] [Google Scholar]
- [50].Shafirovich V, Geacintov NE. Removal of oxidatively generated DNA damage by overlapping repair pathways. Free radical biology & medicine. 2016 doi: 10.1016/j.freeradbiomed.2016.10.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kim Y-J, Wilson DM. Overview of Base Excision Repair Biochemistry. Current molecular pharmacology. 2012;5(1):3–13. doi: 10.2174/1874467211205010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Fortini P, Dogliotti E. Base damage and single-strand break repair: mechanisms and functional significance of short- and long-patch repair subpathways. DNA repair. 2007;6(4):398–409. doi: 10.1016/j.dnarep.2006.10.008. [DOI] [PubMed] [Google Scholar]
- [53].Hashimoto K, Tominaga Y, Nakabeppu Y, Moriya M. Futile short-patch DNA base excision repair of adenine:8-oxoguanine mispair. Nucleic Acids Research. 2004;32(19):5928–5934. doi: 10.1093/nar/gkh909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Petermann E, Keil C, Oei SL. Roles of DNA ligase III and XRCC1 in regulating the switch between short patch and long patch BER. DNA repair. 2006;5(5):544–55. doi: 10.1016/j.dnarep.2005.12.008. [DOI] [PubMed] [Google Scholar]
- [55].Robertson AB, Klungland A, Rognes T, Leiros I. DNA repair in mammalian cells: Base excision repair: the long and short of it. Cellular and molecular life sciences : CMLS. 2009;66(6):981–93. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Boiteux S, Radicella JP. Base excision repair of 8-hydroxyguanine protects DNA from endogenous oxidative stress. Biochimie. 1999;81(1-2):59–67. doi: 10.1016/s0300-9084(99)80039-x. [DOI] [PubMed] [Google Scholar]
- [57].Aburatani H, Hippo Y, Ishida T, Takashima R, Matsuba C, Kodama T, Takao M, Yasui A, Yamamoto K, Asano M. Cloning and characterization of mammalian 8-hydroxyguanine-specific DNA glycosylase/apurinic, apyrimidinic lyase, a functional mutM homologue. Cancer research. 1997;57(11):2151–6. [PubMed] [Google Scholar]
- [58].Arai K, Morishita K, Shinmura K, Kohno T, Kim SR, Nohmi T, Taniwaki M, Ohwada S, Yokota J. Cloning of a human homolog of the yeast OGG1 gene that is involved in the repair of oxidative DNA damage. Oncogene. 1997;14(23):2857–61. doi: 10.1038/sj.onc.1201139. [DOI] [PubMed] [Google Scholar]
- [59].Hirano S, Tominaga Y, Ichinoe A, Ushijima Y, Tsuchimoto D, Honda-Ohnishi Y, Ohtsubo T, Sakumi K, Nakabeppu Y. Mutator phenotype of MUTYH-null mouse embryonic stem cells. The Journal of biological chemistry. 2003;278(40):38121–4. doi: 10.1074/jbc.C300316200. [DOI] [PubMed] [Google Scholar]
- [60].Mo JY, Maki H, Sekiguchi M. Hydrolytic elimination of a mutagenic nucleotide, 8-oxodgtp, by human 18-kilodalton protein - sanitization of nucleotide pool. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(22):11021–11025. doi: 10.1073/pnas.89.22.11021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Rosenquist TA, Zharkov DO, Grollman AP. Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(14):7429–34. doi: 10.1073/pnas.94.14.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Slupska MM, Baikalov C, Luther WM, Chiang JH, Wei YF, Miller JH. Cloning and sequencing a human homolog (hMYH) of the Escherichia coli mutY gene whose function is required for the repair of oxidative DNA damage. J Bacteriol. 1996;178(13):3885–92. doi: 10.1128/jb.178.13.3885-3892.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Fowler RG, White SJ, Koyama C, Moore SC, Dunn RL, Schaaper RM. Interactions among the Escherichia coli mutT, mutM, and mutY damage prevention pathways. DNA repair. 2003;2(2):159–73. doi: 10.1016/s1568-7864(02)00193-3. [DOI] [PubMed] [Google Scholar]
- [64].McAuley-Hecht KE, Leonard GA, Gibson NJ, Thomson JB, Watson WP, Hunter WN, Brown T. Crystal structure of a DNA duplex containing 8-hydroxydeoxyguanine-adenine base pairs. Biochemistry. 1994;33(34):10266–70. doi: 10.1021/bi00200a006. [DOI] [PubMed] [Google Scholar]
- [65].Sakai Y, Furuichi M, Takahashi M, Mishima M, Iwai S, Shirakawa M, Nakabeppu Y. A molecular basis for the selective recognition of 2-hydroxy-dATP and 8-oxo-dGTP by human MTH1. The Journal of biological chemistry. 2002;277(10):8579–87. doi: 10.1074/jbc.M110566200. [DOI] [PubMed] [Google Scholar]
- [66].Schramm VL. Transition States, Analogues, and Drug Development. ACS Chemical Biology. 2013;8(1):71–81. doi: 10.1021/cb300631k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8(3):193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- [68].Rampazzo C, Tozzi MG, Dumontet C, Jordheim LP. The druggability of intracellular nucleotide-degrading enzymes. Cancer chemotherapy and pharmacology. 2016;77(5):883–93. doi: 10.1007/s00280-015-2921-6. [DOI] [PubMed] [Google Scholar]
- [69].Guan Y, Manuel RC, Arvai AS, Parikh SS, Mol CD, Miller JH, Lloyd S, Tainer JA. MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nat Struct Biol. 1998;5(12):1058–64. doi: 10.1038/4168. [DOI] [PubMed] [Google Scholar]
- [70].Bulychev NV, Varaprasad CV, Dormán G, Miller JH, Eisenberg M, Grollman AP, Johnson F. Substrate Specificity of Escherichia coli MutY Protein. Biochemistry. 1996;35(40):13147–13156. doi: 10.1021/bi960694h. [DOI] [PubMed] [Google Scholar]
- [71].Livingston AL, O'Shea VL, Kim T, Kool ET, David SS. Unnatural substrates reveal the importance of 8-oxoguanine for in vivo mismatch repair by MutY. Nature Chemical Biology. 2008;4(1):51–58. doi: 10.1038/nchembio.2007.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Porello SL, Williams SD, Kuhn H, Michaels ML, David SS. Specific recognition of substrate analogs by the DNA mismatch repair enzyme MutY. Journal of the American Chemical Society. 1996;118(44):10684–10692. [Google Scholar]
- [73].Porello SL, Leyes AE, David SS. Single-turnover and pre-steady-state kinetics of the reaction of the adenine glycosylase MutY with mismatch-containing DNA substrates. Biochemistry. 1998;37(42):14756–14764. doi: 10.1021/bi981594+. [DOI] [PubMed] [Google Scholar]
- [74].Chmiel NH, Golinelli MP, Francis AW, David SS. Efficient recognition of substrates and substrate analogs by the adenine glycosylase MutY requires the C-terminal domain. Nucleic Acids Research. 2001;29(2):553–564. doi: 10.1093/nar/29.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Noll DM, Gogos A, Granek JA, Clarke ND. The C-Terminal Domain of the Adenine-DNA Glycosylase MutY Confers Specificity for 8-Oxoguanine·Adenine Mispairs and May Have Evolved from MutT, an 8-Oxo-dGTPase. Biochemistry. 1999;38(20):6374–6379. doi: 10.1021/bi990335x. [DOI] [PubMed] [Google Scholar]
- [76].Volk DE, House PG, Thiviyanathan V, Luxon BA, Zhang S, Lloyd RS, Gorenstein DG. Structural similarities between MutT and the C-terminal domain of MutY. Biochemistry. 2000;39(25):7331–6. doi: 10.1021/bi000416p. [DOI] [PubMed] [Google Scholar]
- [77].Fromme JC, Banerjee A, Huang SJ, Verdine GL. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature. 2004;427(6975):652–6. doi: 10.1038/nature02306. [DOI] [PubMed] [Google Scholar]
- [78].Lee S, Verdine GL. Atomic substitution reveals the structural basis for substrate adenine recognition and removal by adenine DNA glycosylase. Proceedings of the National Academy of Sciences. 2009;106(44):18497–18502. doi: 10.1073/pnas.0902908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Francis AW, David SS. Escherichia coli MutY and Fpg utilize a processive mechanism for target locations. Biochemistry. 2003;42(3):801–810. doi: 10.1021/bi026375+. [DOI] [PubMed] [Google Scholar]
- [80].Friedman JI, Stivers JT. Detection of damaged DNA bases by DNA glycosylase enzymes. Biochemistry. 2010;49(24):4957–67. doi: 10.1021/bi100593a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Li X, Wright PM, Lu AL. The C-terminal domain of MutY glycosylase determines the 7,8-dihydro-8-oxo-guanine specificity and is crucial for mutation avoidance. The Journal of biological chemistry. 2000;275(12):8448–55. doi: 10.1074/jbc.275.12.8448. [DOI] [PubMed] [Google Scholar]
- [82].Brinkmeyer MK, Pope MA, David SS. Catalytic Contributions of Key Residues in the Adenine Glycosylase MutY Revealed by pH-dependent Kinetics and Cellular Repair Assays. Chemistry & Biology. 2012;19(2):276–286. doi: 10.1016/j.chembiol.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Fromme JC, Verdine GL. Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM. Nat Struct Biol. 2002;9(7):544–52. doi: 10.1038/nsb809. [DOI] [PubMed] [Google Scholar]
- [84].Banerjee A, Santos WL, Verdine GL. Structure of a DNA glycosylase searching for lesions. Science. 2006;311(5764):1153–7. doi: 10.1126/science.1120288. [DOI] [PubMed] [Google Scholar]
- [85].Qi Y, Spong MC, Nam K, Banerjee A, Jiralerspong S, Karplus M, Verdine GL. Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme. Nature. 2009;462(7274):762–6. doi: 10.1038/nature08561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Dunn AR, Kad NM, Nelson SR, Warshaw DM, Wallace SS. Single Qdot-labeled glycosylase molecules use a wedge amino acid to probe for lesions while scanning along DNA. Nucleic Acids Res. 2011;39(17):7487–98. doi: 10.1093/nar/gkr459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Livingston AL, Kundu S, Henderson Pozzi M, Anderson DW, David SS. Insight into the roles of tyrosine 82 and glycine 253 in the Escherichia coli adenine glycosylase MutY. Biochemistry. 2005;44(43):14179–90. doi: 10.1021/bi050976u. [DOI] [PubMed] [Google Scholar]
- [88].Raetz AG, Xie YL, Kundu S, Brinkmeyer MK, Chang C, David SS. Cancer-associated variants and a common polymorphism of MUTYH exhibit reduced repair of oxidative DNA damage using a GFP-based assay in mammalian cells. Carcinogenesis. 2012;33(11):2301–2309. doi: 10.1093/carcin/bgs270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Wang L, Lee SJ, Verdine GL. Structural Basis for Avoidance of Promutagenic DNA Repair by MutY Adenine DNA Glycosylase. The Journal of biological chemistry. 2015;290(28):17096–105. doi: 10.1074/jbc.M115.657866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Michelson AZ, Rozenberg A, Tian Y, Sun X, Davis J, Francis AW, O'Shea VL, Halasyam M, Manlove AH, David SS, Lee JK. Gas-phase studies of substrates for the DNA mismatch repair enzyme MutY. J Am Chem Soc. 2012;134(48):19839–50. doi: 10.1021/ja309082k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Williams SD, David SS. Evidence that MutY is a monofunctional glycosylase capable of forming a covalent Schiff base intermediate with substrate DNA. Nucleic Acids Research. 1998;26(22):5123–5133. doi: 10.1093/nar/26.22.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Lu A-L, Tsai-Wu J-J, Cillo J. DNA Determinants and Substrate Specificities of Escherichia coli MutY. Journal of Biological Chemistry. 1995;270(40):23582–23588. doi: 10.1074/jbc.270.40.23582. [DOI] [PubMed] [Google Scholar]
- [93].Tsai-Wu JJ, Liu HF, Lu AL. Escherichia coli MutY protein has both N-glycosylase and apurinic/apyrimidinic endonuclease activities on A.C and A.G mispairs. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(18):8779–83. doi: 10.1073/pnas.89.18.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Manuel RC, Hitomi K, Arvai AS, House PG, Kurtz AJ, Dodson ML, McCullough AK, Tainer JA, Lloyd RS. Reaction Intermediates in the Catalytic Mechanism of Escherichia coli MutY DNA Glycosylase. Journal of Biological Chemistry. 2004;279(45):46930–46939. doi: 10.1074/jbc.M403944200. [DOI] [PubMed] [Google Scholar]
- [95].Williams SD, David SS. Formation of a schiff base intermediate is not required for the adenine glycosylase activity of Escherichia coli MutY. Biochemistry. 1999;38(47):15417–15424. doi: 10.1021/bi992013z. [DOI] [PubMed] [Google Scholar]
- [96].Wright PM, Yu J, Cillo J, Lu A-L. The Active Site of the Escherichia coliMutY DNA Adenine Glycosylase. Journal of Biological Chemistry. 1999;274(41):29011–29018. doi: 10.1074/jbc.274.41.29011. [DOI] [PubMed] [Google Scholar]
- [97].Zharkov DO, Gilboa R, Yagil I, Kycia JH, Gerchman SE, Shoham G, Grollman AP. Role for lysine 142 in the excision of adenine from A:G mispairs by MutY DNA glycosylase of Escherichia coli. Biochemistry. 2000;39(48):14768–78. doi: 10.1021/bi001538k. [DOI] [PubMed] [Google Scholar]
- [98].McCann JAB, Berti PJ. Transition-State Analysis of the DNA Repair Enzyme MutY. Journal of the American Chemical Society. 2008;130(17):5789–5797. doi: 10.1021/ja711363s. [DOI] [PubMed] [Google Scholar]
- [99].Parikh SS, Mol CD, Hosfield DJ, Tainer JA. Envisioning the molecular choreography of DNA base excision repair. Curr Opin Struct Biol. 1999;9(1):37–47. doi: 10.1016/s0959-440x(99)80006-2. [DOI] [PubMed] [Google Scholar]
- [100].Kellie JL, Wilson KA, Wetmore SD. Standard Role for a Conserved Aspartate or More Direct Involvement in Deglycosylation? An ONIOM and MD Investigation of Adenine-DNA Glycosylase. Biochemistry. 2013;52(48):8753–65. doi: 10.1021/bi401310w. [DOI] [PubMed] [Google Scholar]
- [101].Williams SD, David SS. A single engineered point mutation in the adenine glycosylase MutY confers bifunctional glycosylase/AP lyase activity. Biochemistry. 2000;39(33):10098–10109. doi: 10.1021/bi0004652. [DOI] [PubMed] [Google Scholar]
- [102].Davies GJ, Gloster TM, Henrissat B. Recent structural insights into the expanding world of carbohydrate-active enzymes. Curr Opin Struct Biol. 2005;15(6):637–45. doi: 10.1016/j.sbi.2005.10.008. [DOI] [PubMed] [Google Scholar]
- [103].Vocadlo DJ, Davies GJ. Mechanistic insights into glycosidase chemistry. Curr Opin Chem Biol. 2008;12(5):539–55. doi: 10.1016/j.cbpa.2008.05.010. [DOI] [PubMed] [Google Scholar]
- [104].Zechel DL, Withers SG. Glycosidase Mechanisms: Anatomy of a Finely Tuned Catalyst. Accounts of Chemical Research. 2000;33(1):11–18. doi: 10.1021/ar970172+. [DOI] [PubMed] [Google Scholar]
- [105].Zechel DL, Withers SG. Dissection of nucleophilic and acid-base catalysis in glycosidases. Curr Opin Chem Biol. 2001;5(6):643–9. doi: 10.1016/s1367-5931(01)00260-5. [DOI] [PubMed] [Google Scholar]
- [106].McCann JA, Berti PJ. Adenine release is fast in MutY-catalyzed hydrolysis of G:A and 8-Oxo-G:A DNA mismatches. The Journal of biological chemistry. 2003;278(32):29587–92. doi: 10.1074/jbc.M212474200. [DOI] [PubMed] [Google Scholar]
- [107].Parker A, Gu Y, Mahoney W, Lee SH, Singh KK, Lu AL. Human homolog of the MutY repair protein (hMYH) physically interacts with proteins involved in long patch DNA base excision repair. The Journal of biological chemistry. 2001;276(8):5547–55. doi: 10.1074/jbc.M008463200. [DOI] [PubMed] [Google Scholar]
- [108].Yang HJ, Clendenin WM, Wong D, Demple B, Slupska MM, Chiang JH, Miller JH. Enhanced activity of adenine-DNA glycosylase (Myh) by apurinic/apyrimidinic endonuclease (Ape1) in mammalian base excision repair of an A/GO mismatch. Nucleic Acids Research. 2001;29(3):743–752. doi: 10.1093/nar/29.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Pope MA, Porello SL, David SS. Escherichia coli apurinic-apyrimidinic endonucleases enhance the turnover of the adenine glycosylase MutY with G : A substrates. Journal of Biological Chemistry. 2002;277(25):22605–22615. doi: 10.1074/jbc.M203037200. [DOI] [PubMed] [Google Scholar]
- [110].Luncsford PJ, Manvilla BA, Patterson DN, Malik SS, Jin J, Hwang BJ, Gunther R, Kalvakolanu S, Lipinski LJ, Yuan W, Lu W, Drohat AC, Lu AL, Toth EA. Coordination of MYH DNA glycosylase and APE1 endonuclease activities via physical interactions. DNA repair. 2013;12(12):1043–52. doi: 10.1016/j.dnarep.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Pope MA, Chmiel NH, David SS. Insight into the functional consequences of hMYH variants associated with colorectal cancer: distinct differences in the adenine glycosylase activity and the response to AP endonucleases of Y150C and G365D murine MYH. DNA repair. 2005;4(3):315–325. doi: 10.1016/j.dnarep.2004.10.003. [DOI] [PubMed] [Google Scholar]
- [112].Galiatsatos P, Foulkes WD. Familial Adenomatous Polyposis. Am J Gastroenterol. 2006;101(2):385–398. doi: 10.1111/j.1572-0241.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- [113].Moriya M, Grollman AP. Mutations in the mutY gene of Escherichia coli enhance the frequency of targeted G:C-->T:a transversions induced by a single 8-oxoguanine residue in single-stranded DNA. Molecular & general genetics : MGG. 1993;239(1-2):72–6. doi: 10.1007/BF00281603. [DOI] [PubMed] [Google Scholar]
- [114].Thomas D, Scot AD, Barbey R, Padula M, Boiteux S. Inactivation of OGG1 increases the incidence of G . C-->T . A transversions in Saccharomyces cerevisiae: evidence for endogenous oxidative damage to DNA in eukaryotic cells. Molecular & general genetics : MGG. 1997;254(2):171–8. doi: 10.1007/s004380050405. [DOI] [PubMed] [Google Scholar]
- [115].Jones S, Emmerson P, Maynard J, Best JM, Jordan S, Williams GT, Sampson JR, Cheadle JP. Biallelic germline mutations in MYH predispose to multiple colorectal adenoma and somatic G:C-->T:A mutations. Hum Mol Genet. 2002;11(23):2961–7. doi: 10.1093/hmg/11.23.2961. [DOI] [PubMed] [Google Scholar]
- [116].Cheadle JP, Dolwani S, Sampson JR. Inherited defects in the DNA glycosylase MYH cause multiple colorectal adenoma and carcinoma. Carcinogenesis. 2003;24(7):1281–2. doi: 10.1093/carcin/bgg068. author reply 1283. [DOI] [PubMed] [Google Scholar]
- [117].Cheadle JP, Sampson JR. Exposing the MYtH about base excision repair and human inherited disease. Hum Mol Genet. 2003;12:R159–65. doi: 10.1093/hmg/ddg259. Spec No 2. [DOI] [PubMed] [Google Scholar]
- [118].Sampson JR, Dolwani S, Jones S, Eccles D, Ellis A, Evans DG, Frayling I, Jordan S, Maher ER, Mak T, Maynard J, Pigatto F, Shaw J, Cheadle JP. Autosomal recessive colorectal adenomatous polyposis due to inherited mutations of MYH. Lancet. 2003;362(9377):39–41. doi: 10.1016/S0140-6736(03)13805-6. [DOI] [PubMed] [Google Scholar]
- [119].Fleischmann C, Peto J, Cheadle J, Shah B, Sampson J, Houlston RS. Comprehensive analysis of the contribution of germline MYH variation to early-onset colorectal cancer. Int J Cancer. 2004;109(4):554–8. doi: 10.1002/ijc.20020. [DOI] [PubMed] [Google Scholar]
- [120].Cheadle JP, Sampson JR. MUTYH-associated polyposis-From defect in base excision repair to clinical genetic testing. DNA repair. 2007;6(3):274–279. doi: 10.1016/j.dnarep.2006.11.001. [DOI] [PubMed] [Google Scholar]
- [121].Lee S, Uttamapinant C, Verdine GL. A Concise Synthesis of 4‘-Fluoro Nucleosides. Organic Letters. 2007;9(24):5007–5009. doi: 10.1021/ol702222z. [DOI] [PubMed] [Google Scholar]
- [122].Sieber OM, Lipton L, Crabtree M, Heinimann K, Fidalgo P, Phillips RK, Bisgaard ML, Orntoft TF, Aaltonen LA, Hodgson SV, Thomas HJ, Tomlinson IP. Multiple colorectal adenomas, classic adenomatous polyposis, and germ-line mutations in MYH. N Engl J Med. 2003;348(9):791–9. doi: 10.1056/NEJMoa025283. [DOI] [PubMed] [Google Scholar]
- [123].Castillejo A, Vargas G, Castillejo MI, Navarro M, Barbera VM, Gonzalez S, Hernandez-Illan E, Brunet J, Ramon y Cajal T, Balmana J, Oltra S, Iglesias S, Velasco A, Solanes A, Campos O, Sanchez Heras AB, Gallego J, Carrasco E, Gonzalez Juan D, Segura A, Chirivella I, Juan MJ, Tena I, Lazaro C, Blanco I, Pineda M, Capella G, Soto JL. Prevalence of germline MUTYH mutations among Lynch-like syndrome patients. Eur J Cancer. 2014;50(13):2241–50. doi: 10.1016/j.ejca.2014.05.022. [DOI] [PubMed] [Google Scholar]
- [124].Lubbe SJ, Di Bernardo MC, Chandler IP, Houlston RS. Clinical implications of the colorectal cancer risk associated with MUTYH mutation. J Clin Oncol. 2009;27(24):3975–80. doi: 10.1200/JCO.2008.21.6853. [DOI] [PubMed] [Google Scholar]
- [125].Lynch HT, Lynch JF, Attard TA. Diagnosis and management of hereditary colorectal cancer syndromes: Lynch syndrome as a model. CMAJ. 2009;181(5):273–80. doi: 10.1503/cmaj.071574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Koehler U, Grabowski M, Bacher U, Holinski-Feder E. A new interphase fluorescence in situ hybridization approach for genomic rearrangements involving MLH1 and MSH6 in hereditary nonpolyposis colorectal cancer-suspected mutation-negative patients. Cancer genetics and cytogenetics. 2007;175(1):81–4. doi: 10.1016/j.cancergencyto.2007.01.008. [DOI] [PubMed] [Google Scholar]
- [127].Morak M, Koehler U, Schackert HK, Steinke V, Royer-Pokora B, Schulmann K, Kloor M, Hochter W, Weingart J, Keiling C, Massdorf T, Holinski-Feder E. Biallelic MLH1 SNP cDNA expression or constitutional promoter methylation can hide genomic rearrangements causing Lynch syndrome. Journal of medical genetics. 2011;48(8):513–9. doi: 10.1136/jmedgenet-2011-100050. [DOI] [PubMed] [Google Scholar]
- [128].Morak M, Heidenreich B, Keller G, Hampel H, Laner A, de la Chapelle A, Holinski-Feder E. Biallelic MUTYH mutations can mimic Lynch syndrome. Eur J Hum Genet. 2014;22(11):1334–1337. doi: 10.1038/ejhg.2014.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Chmiel NH, Livingston AL, David SS. Insight into the functional consequences of inherited variants of the hMYH adenine glycosylase associated with colorectal cancer: Complementation assays with hMYH variants, and pre-steady-state kinetics of the corresponding mutated E-coli enzymes. Journal of Molecular Biology. 2003;327(2):431–443. doi: 10.1016/s0022-2836(03)00124-4. [DOI] [PubMed] [Google Scholar]
- [130].Ali M, Kim H, Cleary S, Cupples C, Gallinger S, Bristow R. Characterization of mutant MUTYH proteins associated with familial colorectal cancer. Gastroenterology. 2008;135(2):499–507. doi: 10.1053/j.gastro.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Kundu S, Brinkmeyer MK, Livingston AL, David SS. Adenine removal activity and bacterial complementation with the human MutY homologue (MUTYH) and Y165C, G382D, P391L and Q324R variants associated with colorectal cancer. DNA repair. 2009;8(12):1400–10. doi: 10.1016/j.dnarep.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Wooden SH, Bassett HM, Wood TG, McCullough AK. Identification of critical residues required for the mutation avoidance function of human MutY (hMYH) and implications in colorectal cancer. Cancer Lett. 2004;205(1):89–95. doi: 10.1016/j.canlet.2003.10.006. [DOI] [PubMed] [Google Scholar]
- [133].Kundu S, Brinkmeyer MK, Eigenheer RA, David SS. Ser 524 is a phosphorylation site in MUTYH and Ser 524 mutations alter 8-oxoguanine (OG): A mismatch recognition. DNA repair. 2010;9(10):1026–1037. doi: 10.1016/j.dnarep.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Brinkmeyer MK, David SS. Distinct functional consequences of MUTYH variants associated with colorectal cancer: Damaged DNA affinity, glycosylase activity and interaction with PCNA and Hus1. DNA repair. 2015;34:39–51. doi: 10.1016/j.dnarep.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Out AA, Tops CM, Nielsen M, Weiss MM, van Minderhout IJ, Fokkema IF, Buisine MP, Claes K, Colas C, Fodde R, Fostira F, Franken PF, Gaustadnes M, Heinimann K, Hodgson SV, Hogervorst FB, Holinski-Feder E, Lagerstedt-Robinson K, Olschwang S, van den Ouweland AM, Redeker EJ, Scott RJ, Vankeirsbilck B, Gronlund RV, Wijnen JT, Wikman FP, Aretz S, Sampson JR, Devilee P, den Dunnen JT, Hes FJ. Leiden Open Variation Database of the MUTYH gene. Hum Mutat. 2010;31(11):1205–15. doi: 10.1002/humu.21343. [DOI] [PubMed] [Google Scholar]
- [136].Win AK, Jenkins MA, Dowty JG, Antoniou AC, Lee A, Giles GG, Buchanan DD, Clendenning M, Rosty C, Ahnen DJ, Thibodeau SN, Casey G, Gallinger S, Le Marchand L, Haile RW, Potter JD, Zheng Y, Lindor NM, Newcomb PA, Hopper JL, MacInnis RJ. Prevalence and Penetrance of Major Genes and Polygenes for Colorectal Cancer. Cancer Epidemiol Biomarkers Prev. 2016 doi: 10.1158/1055-9965.EPI-16-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Nielsen M, van Steenbergen LN, Jones N, Vogt S, Vasen HF, Morreau H, Aretz S, Sampson JR, Dekkers OM, Janssen-Heijnen ML, Hes FJ. Survival of MUTYH-associated polyposis patients with colorectal cancer and matched control colorectal cancer patients. J Natl Cancer Inst. 2010;102(22):1724–30. doi: 10.1093/jnci/djq370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Kasahara M, Osawa K, Yoshida K, Miyaishi A, Osawa Y, Inoue N, Tsutou A, Tabuchi Y, Tanaka K, Yamamoto M, Shimada E, Takahashi J. Association of MUTYH Gln324His and APEX1 Asp148Glu with colorectal cancer and smoking in a Japanese population. J Exp Clin Cancer Res. 2008;27:49. doi: 10.1186/1756-9966-27-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Miyaishi A, Osawa K, Osawa Y, Inoue N, Yoshida K, Kasahara M, Tsutou A, Tabuchi Y, Sakamoto K, Tsubota N, Takahashi J. MUTYH Gln324His gene polymorphism and genetic susceptibility for lung cancer in a Japanese population. J Exp Clin Cancer Res. 2009;28:10. doi: 10.1186/1756-9966-28-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Picelli S, Zajac P, Zhou XL, Edler D, Lenander C, Dalen J, Hjern F, Lundqvist N, Lindforss U, Pahlman L, Smedh K, Tornqvist A, Holm J, Janson M, Andersson M, Ekelund S, Olsson L, Lundeberg J, Lindblom A. Common variants in human CRC genes as low-risk alleles. Eur J Cancer. 2010;46(6):1041–8. doi: 10.1016/j.ejca.2010.01.013. [DOI] [PubMed] [Google Scholar]
- [142].Tao H, Shinmura K, Suzuki M, Kono S, Mibu R, Tanaka M, Kakeji Y, Maehara Y, Okamura T, Ikejiri K, Futami K, Yasunami Y, Maekawa T, Takenaka K, Ichimiya H, Imaizumi N, Sugimura H. Association between genetic polymorphisms of the base excision repair gene MUTYH and increased colorectal cancer risk in a Japanese population. Cancer Sci. 2008;99(2):355–60. doi: 10.1111/j.1349-7006.2007.00694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Turco E, Ventura I, Minoprio A, Russo MT, Torreri P, Degan P, Molatore S, Ranzani GN, Bignami M, Mazzei F. Understanding the role of the Q338H MUTYH variant in oxidative damage repair. Nucleic Acids Res. 2013;41(7):4093–103. doi: 10.1093/nar/gkt130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Shi G, Chang D-Y, Cheng C-C, Guan X, Venclovas Č, Lu AL. Physical and functional interactions between MutY glycosylase homologue (MYH) and checkpoint proteins Rad9–Rad1–Hus1. Biochemical Journal. 2006;400:53–62. doi: 10.1042/BJ20060774. Pt 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Komine K, Shimodaira H, Takao M, Soeda H, Zhang X, Takahashi M, Ishioka C. Functional Complementation Assay for 47 MUTYH Variants in a MutY-Disrupted Escherichia Coli Strain. Human Mutation. 2015;36(7):704–711. doi: 10.1002/humu.22794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Shi G, Chang DY, Cheng CC, Guan X, Venclovas C, Lu AL. Physical and functional interactions between MutY glycosylase homologue (MYH) and checkpoint proteins Rad9-Rad1-Hus1. Biochem J. 2006;400(1):53–62. doi: 10.1042/BJ20060774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Hayashi H, Tominaga Y, Hirano S, McKenna AE, Nakabeppu Y, Matsumoto Y. Replication-associated repair of adenine:8-oxoguanine mispairs by MYH. Curr Biol. 2002;12(4):335–9. doi: 10.1016/s0960-9822(02)00686-3. [DOI] [PubMed] [Google Scholar]
- [148].Matsumoto Y. Molecular mechanism of PCNA-dependent base excision repair. Prog Nucleic Acid Res Mol Biol. 2001;68:129–38. doi: 10.1016/s0079-6603(01)68095-4. [DOI] [PubMed] [Google Scholar]
- [149].Trasvina-Arenas CH, Lopez-Castillo LM, Sanchez-Sandoval E, Brieba LG. Dispensability of the [4Fe-4S] cluster in novel homologues of adenine glycosylase MutY. The FEBS journal. 2016;283(3):521–40. doi: 10.1111/febs.13608. [DOI] [PubMed] [Google Scholar]
- [150].Chepanoske CL, Golinelli MP, Williams SD, David SS. Positively charged residues within the iron-sulfur cluster loop of E. coli MutY participate in damage recognition and removal. Arch Biochem Biophys. 2000;380(1):11–9. doi: 10.1006/abbi.2000.1890. [DOI] [PubMed] [Google Scholar]
- [151].Golinelli MP, Chmiel NH, David SS. Site-directed mutagenesis of the cysteine ligands to the [4Fe-4S] cluster of Escherichia coli MutY. Biochemistry. 1999;38(22):6997–7007. doi: 10.1021/bi982300n. [DOI] [PubMed] [Google Scholar]
- [152].Lukianova OA, David SS. A role for iron-sulfur clusters in DNA repair. Current Opinion in Chemical Biology. 2005;9(2):145–151. doi: 10.1016/j.cbpa.2005.02.006. [DOI] [PubMed] [Google Scholar]
- [153].Fuss JO, Tsai CL, Ishida JP, Tainer JA. Emerging critical roles of Fe-S clusters in DNA replication and repair. Biochimica et biophysica acta. 2015;1853(6):1253–71. doi: 10.1016/j.bbamcr.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Porello SL, Cannon MJ, David SS. A substrate recognition role for the [4Fe-4S](2+) cluster of the DNA repair glycosylase MutY. Biochemistry. 1998;37(18):6465–6475. doi: 10.1021/bi972433t. [DOI] [PubMed] [Google Scholar]
- [155].Messick TE, Chmiel NH, Golinelli MP, Langer MR, Joshua-Tor L, David SS. Noncysteinyl coordination to the [4Fe-4S](2+) cluster of the DNA repair adenine glycosylase MutY introduced via site-directed mutagenesis. Structural characterization of an unusual histidinyl-coordinated cluster. Biochemistry. 2002;41(12):3931–3942. doi: 10.1021/bi012035x. [DOI] [PubMed] [Google Scholar]
- [156].Bian S, Cowan JA. Protein-bound iron–sulfur centers. Form, function, and assembly. Coordination Chemistry Reviews. 1999;190–192:1049–1066. [Google Scholar]
- [157].Boon EM, Livingston AL, Chmiel NH, David SS, Barton JK. DNA-mediated charge transport for DNA repair. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(22):12543–12547. doi: 10.1073/pnas.2035257100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Boal AK, Yavin E, Barton JK. DNA repair glycosylases with a [4Fe-4S] cluster: a redox cofactor for DNA-mediated charge transport? J Inorg Biochem. 2007;101(11-12):1913–21. doi: 10.1016/j.jinorgbio.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Boal AK, Yavin E, Lukianova OA, O'Shea VL, David SS, Barton JK. DNA-bound redox activity of DNA repair glycosylases containing [4Fe-4S] clusters. Biochemistry. 2005;44(23):8397–8407. doi: 10.1021/bi047494n. [DOI] [PubMed] [Google Scholar]
- [160].Boal AK, Genereux JC, Sontz PA, Gralnick JA, Newman DK, Barton JK. Redox signaling between DNA repair proteins for efficient lesion detection. Proceedings of the National Academy of Sciences. 2009;106(36):15237–15242. doi: 10.1073/pnas.0908059106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Lin JC, Singh RR, Cox DL. Theoretical study of DNA damage recognition via electron transfer from the [4Fe-4S] complex of MutY. Biophys J. 2008;95(7):3259–68. doi: 10.1529/biophysj.108.132183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Grodick MA, Muren NB, Barton JK. DNA charge transport within the cell. Biochemistry. 2015;54(4):962–73. doi: 10.1021/bi501520w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Genereux JC, Barton JK. Mechanisms for DNA charge transport. Chemical reviews. 2010;110(3):1642–62. doi: 10.1021/cr900228f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Al-Tassan N, Eisen T, Maynard J, Bridle H, Shah B, Fleischmann C, Sampson JR, Cheadle JP, Houlston RS. Inherited variants in MYH are unlikely to contribute to the risk of lung carcinoma. Hum Genet. 2004;114 doi: 10.1007/s00439-003-1033-2. [DOI] [PubMed] [Google Scholar]
- [165].Nielsen M, Morreau H, Vasen HF, Hes FJ. MUTYH-associated polyposis (MAP) Crit Rev Oncol Hematol. 2011;79(1):1–16. doi: 10.1016/j.critrevonc.2010.05.011. [DOI] [PubMed] [Google Scholar]
- [166].Goto M, Shinmura K, Nakabeppu Y, Tao H, Yamada H, Tsuneyoshi T, Sugimura H. Adenine DNA glycosylase activity of 14 human MutY homolog (MUTYH) variant proteins found in patients with colorectal polyposis and cancer. Hum Mutat. 2010;31(11):E1861–74. doi: 10.1002/humu.21363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Chepanoske CL, Lukianova OA, Lombard M, Golinelli-Cohen MP, David SS. A residue in MutY important for catalysis identified by photocross-linking and mass spectrometry. Biochemistry. 2004;43(3):651–662. doi: 10.1021/bi035537e. [DOI] [PubMed] [Google Scholar]
- [168].Bai H, Jones S, Guan X, Wilson TM, Sampson JR, Cheadle JP, Lu A-L. Functional characterization of two human MutY homolog (hMYH) missense mutations (R227W and V232F) that lie within the putative hMSH6 binding domain and are associated with hMYH polyposis. Nucleic Acids Research. 2005;33(2):597–604. doi: 10.1093/nar/gki209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Middeldorp A, van Puijenbroek M, Nielsen M, Corver WE, Jordanova ES, ter Haar N, Tops CM, Vasen HF, Lips EH, van Eijk R, Hes FJ, Oosting J, Wijnen J, van Wezel T, Morreau H. High frequency of copy-neutral LOH in MUTYH-associated polyposis carcinomas. J Pathol. 2008;216(1):25–31. doi: 10.1002/path.2375. [DOI] [PubMed] [Google Scholar]
- [170].Nielsen M, Franken PF, Reinards TH, Weiss MM, Wagner A, van der Klift H, Kloosterman S, Houwing-Duistermaat JJ, Aalfs CM, Ausems MG, Brocker-Vriends AH, Gomez Garcia EB, Hoogerbrugge N, Menko FH, Sijmons RH, Verhoef S, Kuipers EJ, Morreau H, Breuning MH, Tops CM, Wijnen JT, Vasen HF, Fodde R, Hes FJ. Multiplicity in polyp count and extracolonic manifestations in 40 Dutch patients with MYH associated polyposis coli (MAP) Journal of medical genetics. 2005;42(9):e54. doi: 10.1136/jmg.2005.033217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Miyaki M, Iijima T, Yamaguchi T, Hishima T, Tamura K, Utsunomiya J, Mori T. Germline mutations of the MYH gene in Japanese patients with multiple colorectal adenomas. Mutat Res. 2005;578(1-2):430–3. doi: 10.1016/j.mrfmmm.2005.01.017. [DOI] [PubMed] [Google Scholar]
- [172].Russell AM, Zhang J, Luz J, Hutter P, Chappuis PO, Berthod CR, Maillet P, Mueller H, Heinimann K. Prevalence of MYH germline mutations in Swiss APC mutation-negative polyposis patients. Int J Cancer. 2006;118(8):1937–40. doi: 10.1002/ijc.21470. [DOI] [PubMed] [Google Scholar]
- [173].Yanaru-Fujisawa R, Matsumoto T, Ushijima Y, Esaki M, Hirahashi M, Gushima M, Yao T, Nakabeppu Y, Iida M. Genomic and functional analyses of MUTYH in Japanese patients with adenomatous polyposis. Clin Genet. 2008;73 doi: 10.1111/j.1399-0004.2008.00998.x. [DOI] [PubMed] [Google Scholar]
- [174].Parker AR, Eshleman JR. Human MutY: gene structure, protein functions and interactions, and role in carcinogenesis. Cellular and molecular life sciences : CMLS. 2003;60(10):2064–83. doi: 10.1007/s00018-003-3053-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Chang DY, Lu AL. Functional interaction of MutY homolog with proliferating cell nuclear antigen in fission yeast, Schizosaccharomyces pombe. The Journal of biological chemistry. 2002;277(14):11853–8. doi: 10.1074/jbc.M111739200. [DOI] [PubMed] [Google Scholar]
- [176].Chang DY, Lu AL. Interaction of checkpoint proteins Hus1/Rad1/Rad9 with DNA base excision repair enzyme MutY homolog in fission yeast, Schizosaccharomyces pombe. The Journal of biological chemistry. 2005;280(1):408–17. doi: 10.1074/jbc.M406800200. [DOI] [PubMed] [Google Scholar]
- [177].Luncsford PJ, Chang D-Y, Shi G, Bernstein J, Madabushi A, Patterson DN, Lu AL, Toth EA. A Structural Hinge in Eukaryotic MutY Homologues Mediates Catalytic Activity and Rad9–Rad1–Hus1 Checkpoint Complex Interactions. Journal of Molecular Biology. 2010;403(3):351–370. doi: 10.1016/j.jmb.2010.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Hahm SH, Park JH, Ko SI, Lee YR, Chung IS, Chung JH, Kang LW, Han YS. Knock-down of human MutY homolog (hMYH) decreases phosphorylation of checkpoint kinase 1 (Chk1) induced by hydroxyurea and UV treatment. BMB Rep. 2011;44(5):352–7. doi: 10.5483/BMBRep.2011.44.5.352. [DOI] [PubMed] [Google Scholar]
- [179].Han SH, Hahm S-H, Tran AHV, Chung JH, Hong M-K, Paik H-D, Kim K-S, Han YS. A physical association between the human mutY homolog (hMYH) and DNA topoisomerase II-binding protein 1 (hTopBP1) regulates Chk1-induced cell cycle arrest in HEK293 cells. Cell & Bioscience. 2015;5(1):1–11. doi: 10.1186/s13578-015-0042-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Olschwang S, Blanche H, de Moncuit C, Thomas G. Similar colorectal cancer risk in patients with monoallelic and biallelic mutations in the MYH gene identified in a population with adenomatous polyposis. Genet Test. 2007;11(3):315–20. doi: 10.1089/gte.2007.9995. [DOI] [PubMed] [Google Scholar]
- [181].Hwang BJ, Jin J, Gao Y, Shi G, Madabushi A, Yan A, Guan X, Zalzman M, Nakajima S, Lan L, Lu AL. SIRT6 protein deacetylase interacts with MYH DNA glycosylase, APE1 endonuclease, and Rad9-Rad1-Hus1 checkpoint clamp. BMC Mol Biol. 2015;16:12. doi: 10.1186/s12867-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, Barnes DE. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(23):13300–5. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Minowa O, Arai T, Hirano M, Monden Y, Nakai S, Fukuda M, Itoh M, Takano H, Hippou Y, Aburatani H, Masumura K, Nohmi T, Nishimura S, Noda T. Mmh/Ogg1 gene inactivation results in accumulation of 8-hydroxyguanine in mice. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(8):4156–61. doi: 10.1073/pnas.050404497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Oka S, Ohno M, Tsuchimoto D, Sakumi K, Furuichi M, Nakabeppu Y. Two distinct pathways of cell death triggered by oxidative damage to nuclear and mitochondrial DNAs. EMBO J. 2008;27(2):421–32. doi: 10.1038/sj.emboj.7601975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Sakamoto K, Tominaga Y, Yamauchi K, Nakatsu Y, Sakumi K, Yoshiyama K, Egashira A, Kura S, Yao T, Tsuneyoshi M, Maki H, Nakabeppu Y, Tsuzuki T. MUTYH-null mice are susceptible to spontaneous and oxidative stress induced intestinal tumorigenesis. Cancer research. 2007;67(14):6599–604. doi: 10.1158/0008-5472.CAN-06-4802. [DOI] [PubMed] [Google Scholar]
- [186].Sakumi K, Tominaga Y, Furuichi M, Xu P, Tsuzuki T, Sekiguchi M, Nakabeppu Y. Ogg1 knockout-associated lung tumorigenesis and its suppression by Mth1 gene disruption. Cancer research. 2003;63(5):902–5. [PubMed] [Google Scholar]
- [187].Sakamoto K, Tominaga Y, Yamauchi K, Nakatsu Y, Sakumi K, Yoshiyama K, Egashira A, Kura S, Yao T, Tsuneyoshi M, Maki H, Nakabeppu Y, Tsuzuki T. MUTYH-Null Mice Are Susceptible to Spontaneous and Oxidative Stress–Induced Intestinal Tumorigenesis. Cancer research. 2007;67(14):6599–6604. doi: 10.1158/0008-5472.CAN-06-4802. [DOI] [PubMed] [Google Scholar]
- [188].Foti JJ, Devadoss B, Winkler JA, Collins JJ, Walker GC. Oxidation of the Guanine Nucleotide Pool Underlies Cell Death by Bactericidal Antibiotics. Science. 2012;336(6079):315–319. doi: 10.1126/science.1219192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Oka S, Leon J, Tsuchimoto D, Sakumi K, Nakabeppu Y. MUTYH, an adenine DNA glycosylase, mediates p53 tumor suppression via PARP-dependent cell death. Oncogenesis. 2014;3:e121. doi: 10.1038/oncsis.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Arai T, Fukae J, Hatano T, Kubo S, Ohtsubo T, Nakabeppu Y, Mori H, Mizuno Y, Hattori N. Up-regulation of hMUTYH, a DNA repair enzyme, in the mitochondria of substantia nigra in Parkinson's disease. Acta Neuropathol. 2006;112(2):139–45. doi: 10.1007/s00401-006-0081-9. [DOI] [PubMed] [Google Scholar]
- [191].Sheng Z, Oka S, Tsuchimoto D, Abolhassani N, Nomaru H, Sakumi K, Yamada H, Nakabeppu Y. 8-Oxoguanine causes neurodegeneration during MUTYH-mediated DNA base excision repair. The Journal of Clinical Investigation. 2012;122(12):4344–4361. doi: 10.1172/JCI65053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].The Nobel Prize in Chemistry 2015 - Popular Information. 2016 http://www.nobelprize.org/nobel_prizes/chemistry/laureates/2015/popular.html. [Google Scholar]