Abstract
Purpose
Tumor-infiltrating lymphocytes (TILs) have prognostic significance in many cancers, yet their roles in glioblastoma (GBM) have not been fully defined. We hypothesized TILs in GBM are associated with molecular alterations, histologies and survival.
Experimental Design
We used data from The Cancer Genome Atlas (TCGA) to investigate molecular, histologic and clinical correlates of TILs in GBMs. Lymphocytes were categorized as absent, present or abundant in histopathologic images from 171 TCGA GBMs. Associations were examined between lymphocytes and histologic features, mutations, copy number alterations, CpG island methylator phenotype, transcriptional class and survival. We validated histologic findings using CD3G gene expression.
Results
We found a positive correlation between TILs and GBMs with gemistocytes, sarcomatous cells, epithelioid cells and giant cells. Lymphocytes were enriched in the mesenchymal transcriptional class and strongly associated with mutations in NF1 and RB1. These mutations are frequent in the mesenchymal class and characteristic of gemistocytic, sarcomatous, epithelioid and giant cell histologies. Conversely, TILs were rare in GBMs with small cells and oligodendroglioma components. Lymphocytes were depleted in the classical transcriptional class and in EGFR-amplified and homozygous PTEN-deleted GBMs. These alterations are characteristic of GBMs with small cells and GBMs of the classical transcriptional class. No association with survival was demonstrated.
Conclusions
TILs were enriched in GBMs of the mesenchymal class, strongly associated with mutations in NF1 and RB1 and typical of histologies characterized by these mutations. Conversely, TILs were depleted in the classical class, EGFR-amplified and homozygous PTEN-deleted tumors and rare in histologies characterized by these alterations.
Keywords: Glioblastoma, Lymphocytes, The Cancer Genome Atlas, Transcriptional class, Mesenchymal, Classical, Neurofibromatosis 1 (NF1), Retinoblastoma 1 (RB1), Epidermal growth factor receptor (EGFR), Tumor protein 53 (TP53), Phosphatase and tensin homolog (PTEN)
INTRODUCTION
Glioblastoma (GBM) is the most common and highest grade astrocytoma (World Heath Organization, grade IV). Despite advances in diagnosis and treatment, GBM remains incurable with an average survival of 15 months.(1) Although characterized by dramatic molecular and histologic heterogeneity, current adjuvant treatments inhibit cell division nonspecifically and are not tailored to specific subsets.(1, 2) Molecular classifications of GBM have revealed distinct subclasses with clinical relevance, opening the way for therapies to be directed at class-specific mechanisms.(3, 4) Immunotherapy, for example, has great potential as a tailored therapy. Already FDA-approved for prostate cancer and metastatic melanoma and in clinical trials for other cancer types, immunotherapy remains promising for patients with GBM, offering high tumor-specificity and long-term tumor surveillance.(5–7)
Tumor-infiltrating lymphocytes (TILs) are present in subsets of GBM, yet their biologic activities and clinical relevance have not been fully explored. The presence of TILs suggests an anti-tumor adaptive immune response and might be expected to impact survival. Lymphocytes are present within the stroma of other cancers, including melanoma, colorectal and ovarian carcinoma, where their presence is associated with improved patient outcomes.(8–17) A recent study suggested TILs have prognostic significance in GBM, yet others have shown lymphocytes may impart a less favorable prognosis.(18–21)
A complex relationship exists between the development of cancer, the immunologic response to it and the immunosuppression it causes. In GBM, tumor-mediated immunosuppression is thought to explain functional deficits in cytotoxic lymphocytes.(22) Regulatory lymphocytes are elevated in patients with GBM and suppress anti-tumor activity by inhibiting secretion of cytotoxic cytokines from effector lymphocytes.(23, 24) Thus, while lymphocytes and other immune cells infiltrate GBM, an immunosuppressive tumor milieu likely prevents successful immune-mediated tumor eradication. Recent clinical trials have focused on augmenting the anti-tumor immune response with tumor vaccines or reversing tumor-mediated immunosuppression.
Molecular alterations in GBM and other cancers may be immunogenic. For example, microsatellite instability and methylation aberrations are independent predictors of lymphocyte density in colorectal cancer.(14) Given its molecular and histologic heterogeneity, subsets of GBM may be more immunogenic and responsive to immunotherapy, yet little is known about the relation between immune cells and the tumor microenvironment or molecular classes in GBM. Therefore, the purpose of this study was to identify molecular and histologic correlates of the immune response in GBM. We used whole-slide digitized images of GBMs from The Cancer Genome Atlas (TCGA) linked to multiplatform molecular data. These may provide insight into the biologic significance of TILs in GBM and impact therapy.
MATERIALS AND METHODS
We performed an integrated molecular and histologic analysis using data from TCGA, which molecularly characterized GBMs across multiple platforms, including single-nucleotide polymorphism (SNP) genotyping, mRNA and microRNA profiling, DNA sequencing and methylation analysis.(2) Clinical data, including treatment and survival, is also available.
Digitized Images used for Morphological Analysis
Permanent section histologic slides from TCGA GBMs were provided by contributing institutions. Slides were scanned and digitized at 20X resolution by a high-throughput digital scanner (Aperio, Inc.) at the TCGA Biospecimen Core Resource located at the International Genomics Consortium (Intgen, Phoenix, AZ). The number of slides available for review ranged from 1–9 per case (median, 3).
Ratings of TILs and other Histopathologic Features
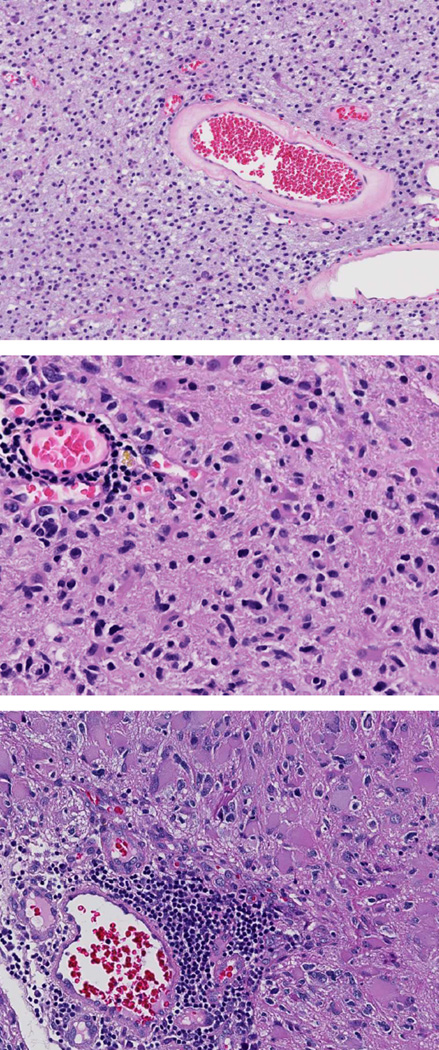
TCGA consortium neuropathologists annotated digitized permanent section histologic slides from 122 TCGA cases for 18 histopathologic features, including inflammation, angiogenesis, necrosis, lymphocytes, common morphologic subtypes and others (Table 1). “Inflammation” as a general category was defined by the presence of inflammatory cells, including lymphocytes, macrophage, neutrophils or their combination. Lymphocytes were identified as small round cells with scant cytoplasm and darkly staining nuclei. Lymphocytes and all other histopathologic features were categorized as absent (0), present (1+) or abundant (2+) by two neuropathologists (MC, KA, RM, NL, RM, MJS, DJB) and adjudicated by a third. Cases with a complete absence of lymphocytes were labeled 0 (absent). Cases with lymphocytes in <50% of tumor tissue were categorized 1+ (present), while cases with lymphocytes in ≥50% were categorized 2+ (abundant) (Figure 1). In addition to these 122 cases, an additional 49 TCGA cases from Emory University Hospital and Henry Ford Health System were annotated for the same 18 histopathologic features using the same criteria by two TCGA neuropathologists (DJB, MJS). All digitized histologic sections and TCGA neuropathology ratings were obtained from the TCGA portal (http://tcga-data.nci.nih.gov.proxy.library.emory.edu/tcga/tcgaHome2.jsp; last accessed November 28, 2012). Digitized images and corresponding neuropathology ratings were the primary source of data.
Table 1.
18 histopathologic features were categorized as absent (0), present (1+), or abundant (2+) in 171 TCGA GBMs. TILs are associated with specific histopathologic features in GBM.
| Histopathologic Feature | Mantel-Haenzel chi-square p-value |
Spearman correlation |
|---|---|---|
| Inflammation | <0.001 | 0.93 |
| Pseudopalisading necrosis | 0.01 | −0.19 |
| Zonal necrosis | 0.04 | 0.16 |
| Neutrophils | 0.09 | 0.13 |
| Macrophages | <0.001 | 0.32 |
| Microvascular hyperplasia | 0.59 | −0.04 |
| Endothelial hyperplasia | 0.05 | −0.15 |
| Epithelial metaplasia | 0.04 | 0.16 |
| Gemistocytes | 0.01 | 0.22 |
| Giant cells | 0.02 | 0.18 |
| Oligodendroglial cells | 0.01 | −0.19 |
| Sarcomatous metaplasia | <0.01 | 0.22 |
| Small cells | <0.01 | −0.24 |
| Mineralization | 0.68 | 0.03 |
| Satellitosis | 0.20 | −0.10 |
| White matter invasion | 0.09 | −0.13 |
| Cortex invasion | 0.71 | −0.03 |
Mantel-Haenzel chi-square test and Spearman correlation were used to examine associations between lymphocytes (0, 1+, 2+) and other histopathologic features (0, 1+, 2+)
Mantel-Haenzel exact chi-square test was used when 25% of the cells had <5 observations
Effective Sample Size 171
Figure 1.
Representative examples of permanent section histologic slides from TCGA cases. Lymphocytes were identified as small round cells with scant cytoplasm and darkly staining nuclei. Cases with a complete absence of lymphocytes were labeled 0 (absent) (top). Cases with lymphocytes present in <50% of tumor tissue were categorized 1+ (present) (middle).Cases with lymphocytes present in ≥50% were categorized 2+ (abundant) (bottom).
Mutations, Copy Number Alterations, Methylation Status and Transcriptional Class
We obtained TCGA mutation and copy number data from the Memorial Sloan Kettering Cancer Genomics Portal (http://www.cbioportal.org/public-portal; last accessed November 28, 2012).(25) Mutation data from whole exome sequencing (NextGen) was obtained for 99 cases. Putative copy-number calls determined with GISTIC 2.0 were obtained for 153 cases. CpG island methylator phenotype (G-CIMP) status was available for 124 cases. Transcriptional class labels for the Verhaak classification were obtained for 162 cases from the TCGA Advanced Working Group. The updated Verhaak labeling extends the original labeled set presented in Verhaak et al. by using the originally labeled samples along with Affymetrix HT_HG-U133A data to label previously unclassified samples.(3)
Validation Dataset
CD3G is the gene that encodes the T-cell surface marker CD3. We used CD3G expression data from TCGA as a marker of lymphocytes to validate our histologic findings in the same set of tumors. CD3G expression data was obtained from the TCGA portal (http://tcga-data.nci.nih.gov.proxy.library.emory.edu/tcga/tcgaHome2.jsp; last accessed February 1, 2012).
Statistical Analyses
Associations between lymphocytes and other histopathologic features were assessed using the Mantel-Haenzel chi-square and exact test and Spearman correlation for ordinal comparisons (0, 1+, 2+ vs 0, 1+, 2+). The Mantel-Haenzel exact test was used when the cell count was <5 in 25% or more of cells. Chi-square and Fisher’s exact test were used for all dichotomous comparisons (0 vs 1+, 2+ combined or 0, 1+ combined vs 2+). We combined categories to identify associations driven by cases with either complete absence or abundance of lymphocytes. The association of lymphocytes with mutations, copy number alterations and G-CIMP status were also assessed using chi-square and Fisher’s exact test. Fisher’s exact test was used when the cell count was <5 in 25% or more of cells. Associations between lymphocytes and transcriptional class were examined using chi-square and Fisher’s exact test. The Spearman correlation was used to measure the correlation between lymphocytes and CD3G expression. Wilcoxon two-sample or Kruskal-Wallis tests were used to detect differences in CD3G expression according to lymphocytes, mutation or copy number status and transcriptional class.
Survival Analysis
Clinical data was obtained from the TCGA data portal. Survival was taken as “days to death” for uncensored patients and “days to last follow-up” for right-censored patients. The association between TILs and survival was examined using the log-rank test.
All p-values reported are two-sided and regarded as statistically significant if p<0.05. The software used for statistical analysis was SAS Version 9.3 (SAS Institute Inc., Cary NC).
RESULTS
TILs are differentially distributed in GBM
Within the 171 GBMs reviewed for this study, tumor-infiltrating lymphocytes (TILs) were absent (0) in 93 cases (54%), present (1+) in 59 cases (35%) and abundant (2+) in 19 cases (11%). There was no significant association between the number of slides analyzed for each case and the level of lymphocytes annotated.
TILs are associated with specific histopathologic features in GBM
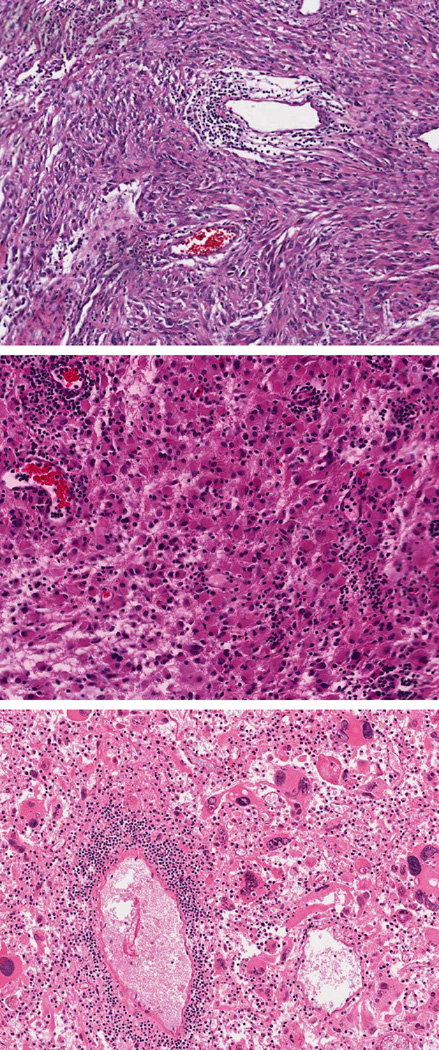
GBMs show a tremendous degree of histologic variability and numerous morphologic subtypes have been recognized, including fibrillary, gemistocytic, epitheliod, small cell, giant cell, gliosarcoma and GBM with oligodendroglioma component. To determine if TILs correlated with specific GBM morphologies or other histopathologic features, we examined their associations using the Mantel-Haenzel chi-square test and Spearman correlation (Table 1). All variables, including lymphocytes and other histopathologic features were categorized as 0, 1+ or 2+. We detected a strong positive correlation between TILs and specific tumor cell morphologies that included gemistocytes, sarcomatous cells, epithelioid cells and giant cells (all p<0.05) (Figure 2). Conversely, TILs were depleted in GBMs characterized by small cells and oligodendroglial cells (both p<0.05). Among other features analyzed, TILs were most tightly correlated with the findings of inflammation (as a general category) and macrophages (both p<0.05). TILs were also statistically associated with both forms of necrosis annotated (pseudopalisading and zonal) (both p<0.05).
Figure 2.
Representative examples of GBM morphologic subtypes associated with the presence of lymphocytes in TCGA permanent section histologic slides. TILs are enriched in GBMs with sarcomatous cells (top), gemistocytes (middle) and giant cells (bottom).
TILs are associated with specific mutations in GBM
In the initial TCGA analysis of GBM, eight genes were identified as significantly mutated, including TP53, PTEN, NF1, EGFR, ERBB2, RB1, PIK3R1 and PIK3CA (genes attaining a false discovery rate <0.1).(2) A subsequent, unbiased genomic analysis identified recurrent mutations in isocitrate dehydrogenase 1 (IDH1).(26) We examined associations between lymphocytes (0, 1+, 2+) and mutations (mutant vs wild type) for these genes using chi-square and Fisher’s exact test (Table 2). Out data set contained no cases with ERBB2 mutations.
Table 2.
TILs are associated with specific mutations in GBM
| Gene | n | Chi square p-value (0, 1+, 2+) |
Chi square p-value (0 vs 1+, 2+ combined) |
Chi square p-value (0, 1+ combined vs 2+) |
|---|---|---|---|---|
| TP53 | 33 | 0.13 | 0.06 | 0.15 |
| PTEN | 29 | 0.15 | 0.06 | 0.44 |
| EGFR | 26 | 0.82 | 0.54 | 1.00 |
| NF1 | 16 | 0.03 | 0.74 | 0.04 |
| RB1 | 10 | 0.04 | 0.04 | 0.22 |
| PIK3R1 | 9 | 0.67 | 0.49 | 1.00 |
| PIK3CA | 7 | 0.72 | 1.00 | 0.50 |
| IDH1 | 4 | 0.53 | 1.00 | 0.32 |
| ERBB2 | 0 | - | - | - |
Chi-square test was used to examine associations between lymphocytes (0, 1+, 2+) and mutations (0, 1)
Fisher’s exact test was used when 25% of the cells had <5 observations
Effective Sample Size 99
Categories were combined to identify associations driven by cases with either complete absence or abundance of lymphocytes
Among cases with mutational data, lymphocytes were absent (0) in 52, present (1+) in 38 and abundant (2+) in 9. We found lymphocytes were strongly associated with mutations in neurofibromatosis 1 (NF1) and retinoblastoma 1 (RB1) (both p<0.05). Nine cases with absent (0) lymphocytes were NF1-mutant (9/52, 17%), while 44% of cases with abundant (2+) lymphocytes harbored mutations in NF1 (4/9). Two cases with absent (0) lymphocytes harbored mutations in RB1 (2/52, 4%). Of cases with lymphocytes present (1+), six were RB1 mutants (6/32, 16%). Two cases with abundant (2+) lymphocytes harbored mutations in RB1 (2/9, 22%).
There was a trend towards significance for TP53 mutations (p=0.12), particularly when cases with present (1+) and abundant (2+) lymphocytes were combined (p=0.06). 25% of cases with absent (0) lymphocytes were TP53-mutant (13/52), while 43% of cases with present (1+) or abundant (2+) lymphocytes harbored mutations in TP53 (20/47).
Since mutations in TP53 are characteristic of both the proneural and mesenchymal transcriptional class, we investigated if mutations were overrepresented in one of these two transcriptional classes.(3) 20 cases harbored mutations in TP53 and had present (1+) or abundant (2+) lymphocytes. Nine (45%) belonged to the mesenchymal transcriptional class and five (25%) belonged to the proneural class (p>0.05).
TILs are associated with specific copy number alterations in GBM
Significant copy number alterations (CNAs) were also identified in the initial TCGA global analysis publication, including amplifications of EGFR, CDK4, PDGFRA, MDM2, MDM4, MET, CDK6, MYCN, CCND2, PIK3CA and AKT3 and deletions of CDKN2A/B, PTEN, CDKN2C, RB1, PARK2 and NF1.(2) We examined the association between TILs and these recurrent amplifications and deletions using chi-square and Fisher’s exact test (Table 3). Lymphocytes were depleted in EGFR-amplified tumors (p<0.05). Amplification of EGFR was present in 65% of cases with absent (0) lymphocytes (53/82) compared to 48% of cases with present (1+) or abundant (2+) lymphocytes. Only 35% of cases with abundant (2+) lymphocytes were EGFR-amplified.
Table 3.
TILs are depleted in EGFR-amplified and homozygous PTEN-deleted tumors
| CNA | (%) | Chi square p-value (0, 1+, 2+) |
Chi square p-value (0 vs 1+, 2+ combined) |
Chi square p-value (0, 1+ combined vs 2+) |
|---|---|---|---|---|
| Amplifications | ||||
| EGFR | 49.3 | 0.05 | 0.04 | 0.06 |
| CDK4 | 14.5 | 0.37 | 0.22 | 0.47 |
| PDGFRA | 14.1 | 0.61 | 0.73 | 0.47 |
| MDM4 | 9.9 | 0.23 | 0.24 | 0.64 |
| MDM2 | 9.3 | 0.81 | 0.82 | 1.00 |
| MET | 8.8 | 0.57 | 0.79 | 0.48 |
| CDK6 | 7.0 | 0.75 | 0.65 | 0.69 |
| CCND2 | 4.0 | 1.00 | 1.00 | 1.00 |
| AKT3 | 3.2 | 1.00 | 1.00 | 1.00 |
| MYCN | 2.6 | 1.00 | 0.62 | 1.00 |
| PIK3CA | 2.4 | 0.69 | 0.60 | 1.00 |
| Deletions | ||||
| CDKN2A | 62.0 | 0.23 | 0.09 | 0.48 |
| CDKN2B | 61.0 | 0.23 | 0.09 | 0.48 |
| PTEN | 10.3 | 0.09 | 0.03 | 0.69 |
| RB1 | 3.6 | 0.87 | 1.00 | 0.57 |
| CDKN2C | 3.6 | 0.71 | 1.00 | 0.51 |
| PARK2 | 2.0 | 1.00 | 1.00 | 1.00 |
| NF1 | 1.2 | 0.07 | 0.10 | 0.30 |
Chi-square test was used to examine associations between lymphocytes (0, 1+, 2+) and CNAs
Fisher’s exact test was used when 25% of the cells had <5 observations
Effective Sample Size 153
Categories were combined to identify associations driven by cases with either complete absence or abundance of lymphocytes
TILs were also depleted in tumors with homozygous deletions of PTEN (p<0.05). 78% of cases with homozygous PTEN-deletion (14/18) had absent (0) lymphocytes. 17% of PTEN-deleted cases had present (1+) lymphocytes (3/18), while only 5% of cases had abundant (2+) lymphocytes (1/18). There was a trend towards significance for NF1 deletions (p=0.07). No other associations between TILs and copy number alterations were noted.
TILs are not associated with the CpG island methylator phenotype
G-CIMP positive tumors represent less than 10% of GBMs, but have significantly improved survival compared to G-CIMP negative tumors.(27) IDH1 mutations have been shown to establish the G-CIMP phenotype.(28) We examined the association between TILs and the CpG island methylator phenotype using chi-square and Fisher’s exact test (data not shown). TILs were not associated with G-CIMP status (p>0.05).
TILs are related to transcriptional class
Verhaak et al. identified four transcriptional classes of GBM using TCGA gene expression data: proneural, neural, classical and mesenchymal.(3) To determine if there was an association between transcriptional class and TILs, we examined each class for its distribution of lymphocytes using chi-square and Fisher’s exact test (Table 4). We found TILs were strongly associated with transcriptional class (p<0.05).
Table 4.
TILs are enriched in the mesenchymal class and depleted in the classical class
| Lymphocytes | Transcriptional Class | ||||
|---|---|---|---|---|---|
| Classical | Mesenchymal | Neural | Proneural | Total | |
| 0 | 31 | 27 | 13 | 19 | 90 |
| 1+ | 10 | 18 | 9 | 18 | 55 |
| 2+ | 1 | 12 | 2 | 2 | 17 |
| Total | 42 | 57 | 24 | 39 | 162 |
Chi-square test was used to examine associations between lymphocytes (0, 1+, 2+) and transcriptional class
Fisher’s exact test was used when 25% of the cells had <5 observations
Effective sample size 162
TILs were enriched in the mesenchymal class compared to all other classes combined (p<0.05). 42% of cases (30/72) with a lymphocytic infiltrate (1+ or 2+) belonged to the mesenchymal class. However, when classical tumors were excluded, there was no statistically significant enrichment of TILs in the mesenchymal class compared to the proneural and neural classes (p>0.05). Cases with abundant lymphocytes were heavily enriched in mesenchymal transcriptional class. 71% of cases (12/17) with abundant lymphocytes (2+) belonged to the mesenchymal class, whereas 12% were proneural, 12% neural and 6% were classical, representing statistically significant enrichment (p<0.05). When classical cases were excluded, there was a still a strong trend towards statistically significant enrichment of abundant (2+) TILs in the mesenchymal class compared to the proneural and neural classes (p=0.07).
Conversely, TILs were depleted in the classical transcriptional case compared to all other classes combined (p<0.05). 74% of classical cases (31/42) were characterized by absent (0) lymphocytes, representing statistically significant depletion (p<0.05). Only one case with abundant (2+) lymphocytes belonged to the classical transcriptional class (1/17). When mesenchymal tumors were excluded, there was still a trend towards statistically significant depletion of TILs (1+ and 2+) in the classical class compared to the proneural and neural classes (p=0.053).
CD3G gene expression is positively correlated with lymphocytes, mutations in TP53, RB1 and the mesenchymal transcriptional class and negatively correlated with EGFR amplification, PTEN deletion and the classical transcriptional class
We used CD3G expression data to validate findings uncovered by our morphologic analysis. There was a positive correlation (0.30) between the histologic categorization of lymphocytes and CD3G expression (p<0.001). GBMs with higher levels of TILs had significantly higher CD3G expression (p<0.001). Those with absent (0), present (1+) and abundant (2+) lymphocytes had CD3G expression of 15.34 (Standard deviation (SD) ± 7.81), 17.55 (SD ± 10.99) and 20.23 (SD ± 13.51), respectively
We examined levels of CD3G expression in cases harboring mutations in RB1, TP53 and NF1. Cases with RB1 mutations tended to have higher levels of CD3G expression than wild type cases (19.1 vs 16.2). Cases with TP53 mutations also showed higher levels of CD3G expression than wild type (17.4 vs 16.0). Cases with NF1 mutations had lower levels of CD3G expression compared to wild type (15.6 vs 16.5) (all p>0.05).
We also investigated CD3G expression in EGFR-amplified and PTEN-deleted cases. EGFR-amplified cases had lower levels of CD3G expression than wild type (15.6 vs 16.7). Likewise, PTEN-deleted cases also had lower levels of CD3G expression (15.5 vs 16.3) (both p>0.05).
Finally, we analyzed CD3G expression levels by transcriptional class. There was a statistically significant difference in expression according to transcriptional class (p<0.001). Cases belonging to the mesenchymal transcriptional class had the highest CD3G expression (19.6), which was significantly higher than that of other classes combined (15.1) (p<0.001). Conversely, CD3G expression was significantly lower in the classical transcriptional class compared to all other classes (14.8 vs 17.1) (p<0.001).
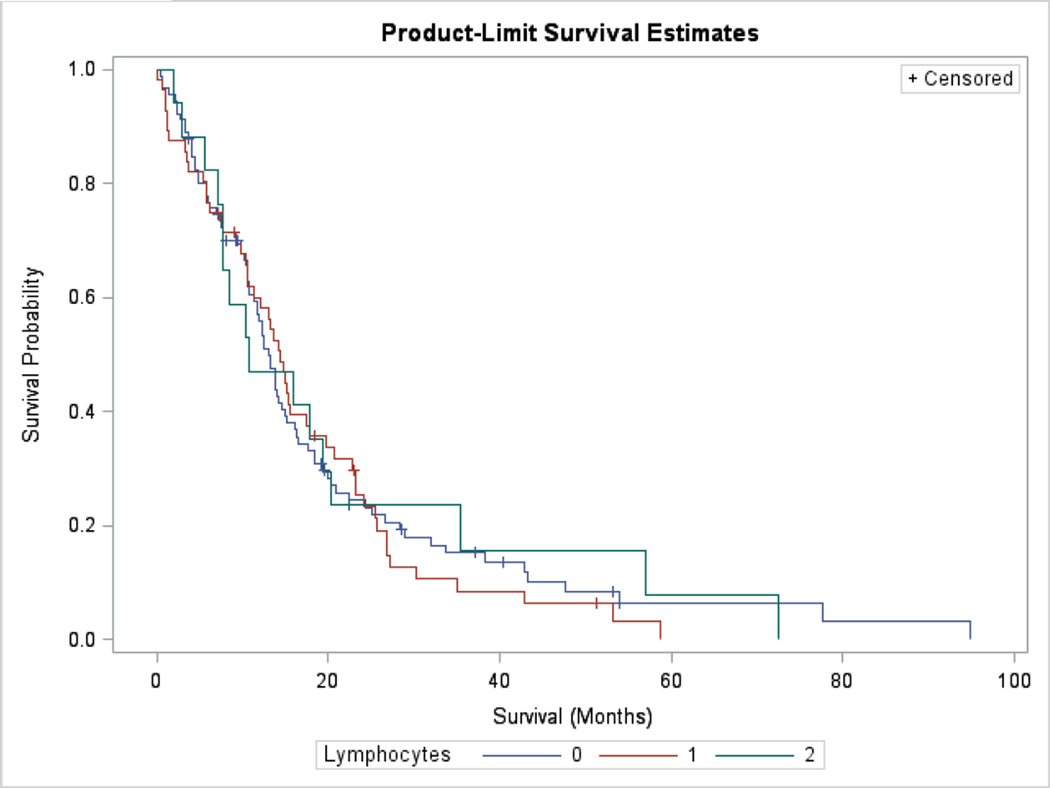
TILs are not associated with prolonged with survival
TILs were not associated with prolonged survival in univariate analyses. We compared the survival of cases with absent (0) lymphocytes, present (1+) and abundant (2+) lymphocytes using a log-rank test (p>0.05) (Figure 3). We found patient age was a highly significant predictor of survival (p<0.001); however, lymphocytes did not vary according to age. The mean age of patients with absent (0), present (1+) and abundant (2+) lymphocytes was 56.9 (SD ± 14.6), 57.3 (SD ± 12.1) and 54.8 years (SD ± 13.1), respectively (p>0.05). TILs were not a significant predictor of survival in an age-adjusted Cox proportional hazards model (data not shown) (p>0.05). If tumors belonging to the classical transcriptional class were excluded, there were no survival differences according to TILs among proneural, neural and mesenchymal tumors (p>0.05). Similarly, if mesenchymal tumors were excluded, there were no survival differences according to TILs among proneural, neural and classical tumors (p>0.05).
Figure 3.
Kaplan–Meier estimates of survival according to absent (0), present (1+) or abundant (2+) lymphocytes. TILs are not associated with improved survival (log-rank p>0.05).
DISCUSSION
Lymphocytes are present in the stroma of many human cancers. The histologic finding of tumor-infiltrating lymphocytes (TILs) is often associated with prolonged survival. Tumors with TILs may have distinctive clinicopathologic features or underlying genetic alterations, which could be relevant for future tailored therapies.
Although prognostically significant in other cancers, the clinical relevance and genetic associations of TILs in GBM remains unclear. The TCGA data set offers an opportunity to study the relationship between morphologic features, molecular alterations and survival in GBM.(3, 4) Verhaak et al. used TGCA gene expression profiles to identify four transcriptional classes, while independent studies of genome methylation uncovered a hypermethylated, G-CIMP+ subset characterized by IDH mutations and overproduction of the oncometabolite 2-hydroxyglutarate (2-HG).(3, 26, 29) Recent evidence suggests GBMs within the mesenchymal transcriptional class have a better response to immunotherapy, suggesting this class may be more immunogenic.(30) We hypothesized TILs are differentially distributed within specific morphologic and molecular classes of GBM.
We found TILs were not uniformly distributed among GBMs, suggesting some are more capable of eliciting an immune response. Indeed, lymphocytes were absent in over half, as determined morphologically by a panel of TCGA neuropathologists. TILs were strongly enriched in the mesenchymal transcriptional class of GBM, which suggests tumors belonging to this subtype are more immunogenic. Seventy-one percent of tumors with abundant (2+) lymphocytes belonged to the mesenchymal class. No other transcriptional class had more than 12% with abundant (2+) TILs, indicating a fundamental difference in transcriptional class with regard to TILs.
Variation of TILs could potentially be accounted for by the extent of necrosis, since the mesenchymal class signature is heavily influenced by the degree of necrosis and the presence of lymphocytes was associated with necrosis in this study.(31) However, mesenchymal GBMs with abundant (2+) TILs did not have substantially different levels of necrosis than those with no TILs. Neither zonal or pseudopalisading necrosis were associated with transcriptional class. Moreover, molecular alterations associated with the mesenchymal class are more likely to be responsible for the association with TILs. Lymphocytes were strongly associated with mutations in NF1 and RB1 and a trend was noted for TP53 mutations. Mutations of NF1 and RB1 are characteristic of the mesenchymal transcriptional class and TP53 mutations are common in both the mesenchymal and proneural subtypes. Interestingly, we found TP53 mutant tumors with abundant (2+) TILs were more common in the mesenchymal than the proneural class, but this did not reach statistical significance.
We also found TILs were more common in certain morphologic subsets of GBM, including those with sarcomatous, giant cell, epithelioid and gemistocytic components. This was of interest since prior studies have shown gliosarcoma, giant cell glioblastoma and gemistocytic astrocytomas are all characterized by a high frequency of TP53 mutations.(32–34) Our own studies using TCGA data indicated sarcomatous components in GBM were associated with NF1, RB1 and TP53 mutations; giant cells were associated with TP53 and RB1 mutations; and epithelioid cells were associated with NF1 and RB1 mutations (data not shown). We did not find an association between TP53 mutations and gemistocytic cells, likely because of inclusion of tumors with low levels of this histologic finding as compared to prior studies, which included morphologically pure gemistocytic astrocytomas of lower grade. Nonetheless, we found the presence of TILs was strongly associated with the mesenchymal transcriptional class as well as the mutations and tumor morphologies associated with this gene signature.
We also found TILs were rare in tumors of the classical transcriptional class, suggesting these may exclude lymphocytes and prevent immune-mediated tumor eradication. Only one case with abundant (2+) lymphocytes belonged to the classical transcriptional class (1/17). We also noted TILs were depleted in EGFR-amplified GBMs, which are frequent in the classical transcriptional class of GBMs, but less common in other classes, including mesenchymal. Since small cell GBMs have been shown to have a high frequency EGFR amplification, we were also encouraged that the histologic presence of small cells within GBMs from the TCGA data set was associated with TIL depletion.(35) Recent evidence suggests EGFR activation may repress the adaptive immune response, potentially through attenuation of MHCI and MHCII expression, and therefore explain the relation between EGFR amplification and lymphocyte depletion.(36) We also found TILs were depleted in PTEN-deleted tumors (homozygous). Loss of PTEN has been shown to increase expression of the immunosuppressive protein B7 homolog 1 (B7-H1), and therefore may account for the association between homozygous PTEN deletion and lymphocyte depletion.(22)
We used CD3G expression data from TCGA to validate our morphologic findings. The CD3G gene encodes a protein that forms the T cell receptor-CD3 complex and is highly specific to T lymphocytes and present in all subsets. The correlation between lymphocytes and CD3G expression was highly statistically significant (p<0.001), however the strength of the correlation was moderate (0.3). Although the tissue used for molecular analysis by TCGA was from the same neoplasm as the tissue used to create slide images, the distribution of TILs in tumors can be irregular and may contribute to a weak correlation. In addition, the detection and evaluation of TILs by molecular and pathologic methods differ considerably and may also lead to a weakened correlation. However, we were encouraged that mutations associated with TILs (RB1 and TP53) tended to have higher levels of CD3G expression, whereas copy number alterations associated with depletion of lymphocytes (EGFR amplification and homozygous PTEN deletion) had lower CD3G expression. Tumors of the mesenchymal transcriptional class had significantly higher CD3G expression than all other subtypes, while those of the classical subtype had significantly lower expression, which corroborates the associations uncovered in our morphologic analysis.
GBMs annotated as having zonal necrosis by TCGA neuropathologists did not have significantly higher levels of CD3G expression and those with pseudopalisading necrosis had lower levels of CD3G expression, suggesting TILs may indeed represent an anti-tumor adaptive immune response rather than a response to necrosis.
In summary, our analysis of digitized, whole slide H&E-stained images from TCGA had the advantage of a large number of cases and high quality, multi-platform molecular analysis. It illustrates the power of networks such as the TCGA to link multi-platform molecular analysis to histopathologic findings in cancer with implications for future therapy. The categorical classification of lymphocytes as 0, 1+ and 2+ was not optimal since statistical associations with molecular and clinical variables were not as strong. We have not adjusted for multiple comparisons since the purpose of the analysis was to identify common molecular alterations in GBM related to the immune response for future tissue-based analyses. However, our analysis was limited to alterations known as significant and recurrent events in GBM pathogenesis. Furthermore, we are encouraged that each statistically significant association has a biologic rationale. Finally, this classification does not account for the functional activity of lymphocytes or specific lymphocyte subsets. While effector lymphocytes are thought to mediate the anti-tumor response, regulatory lymphocytes may suppress the cytotoxic activity of effector lymphocytes. Thus, effector and regulatory lymphocytes may have distinct molecular and histologic associations. Future studies that examine the molecular correlates of lymphocyte subsets will add substantially to the field of immunotherapy.
Statement of Translational Relevance.
Tumor-infiltrating lymphocytes (TILs) are present in a subset of GBMs, suggesting some are more capable of eliciting an adaptive immune response. We hypothesized TILs are differentially distributed among specific molecular classes of GBM and used histologic data linked to multiplatform molecular analysis from The Cancer Genome Atlas (TCGA) to identify correlates. We have shown that TILs are associated with specific genomic alterations and related to transcriptional class, which may provide insight into the biologic significance of TILs in GBM and predict response to immunotherapy.
ACKNOWLEDGEMENTS
The authors would like to thank the Research Pathology Lab of the Cancer Tissue and Pathology Shared Resource within the Winship Cancer Institute for their assistance with whole slide scanning and digital pathology, especially Candace Chisolm. The authors would also like to thank the leadership and support staff of the TCGA Working Group and Expert Pathology Committee for their assistance in the generation of publicly available data.
FUNDING
This work was supported by US Public Health Service grants from the Clinical and Translational Science Award program, NIH, NCRR UL RR025008, TL1 RR025010; the In Silico Center for Brain Tumor Research Contract ST12-1100 (NCI-SAIC Frederick); and the Winship Cancer Center Support Grant (P30 CA138292).
Footnotes
Conflict of interest: The authors have no conflicts of interest.
REFERENCES
- 1.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 5.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kantoff PW, Higano CS, Shore ND, Berger ER, Small EJ, Penson DF, et al. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N Engl J Med. 2010;363:411–422. doi: 10.1056/NEJMoa1001294. [DOI] [PubMed] [Google Scholar]
- 7.Heimberger AB, Sampson JH. Immunotherapy coming of age: what will it take to make it standard of care for glioblastoma? Neuro Oncol. 2011;13:3–13. doi: 10.1093/neuonc/noq169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark WH, Jr., Elder DE, Guerry Dt, Braitman LE, Trock BJ, Schultz D, et al. Model predicting survival in stage I melanoma based on tumor progression. J Natl Cancer Inst. 1989;81:1893–1904. doi: 10.1093/jnci/81.24.1893. [DOI] [PubMed] [Google Scholar]
- 9.Clemente CG, Mihm MC, Jr, Bufalino R, Zurrida S, Collini P, Cascinelli N. Prognostic value of tumor infiltrating lymphocytes in the vertical growth phase of primary cutaneous melanoma. Cancer. 1996;77:1303–1310. doi: 10.1002/(SICI)1097-0142(19960401)77:7<1303::AID-CNCR12>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 10.Dunn GP, Dunn IF, Curry WT. Focus on TILs: Prognostic significance of tumor infiltrating lymphocytes in human glioma. Cancer Immun. 2007;7:12. [PMC free article] [PubMed] [Google Scholar]
- 11.Laghi L, Bianchi P, Grizzi F, Malesci A. How dense, how intense? Role of tumour-infiltrating lymphocytes across colorectal cancer stages. Re: Nosho et al. Tumour-infiltrating T-cell subsets, molecular changes in colorectal cancer, and prognosis: cohort study and literature review. J Pathol. 2010;222:350–366. doi: 10.1002/path.2774. J Pathol. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menon AG, Janssen-van Rhijn CM, Morreau H, Putter H, Tollenaar RA, van de Velde CJ, et al. Immune system and prognosis in colorectal cancer: a detailed immunohistochemical analysis. Lab Invest. 2004;84:493–501. doi: 10.1038/labinvest.3700055. [DOI] [PubMed] [Google Scholar]
- 13.Ropponen KM, Eskelinen MJ, Lipponen PK, Alhava E, Kosma VM. Prognostic value of tumour-infiltrating lymphocytes (TILs) in colorectal cancer. J Pathol. 1997;182:318–324. doi: 10.1002/(SICI)1096-9896(199707)182:3<318::AID-PATH862>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 14.Nosho K, Baba Y, Tanaka N, Shima K, Hayashi M, Meyerhardt JA, et al. Tumour-infiltrating T-cell subsets, molecular changes in colorectal cancer, and prognosis: cohort study and literature review. J Pathol. 2010;222:350–366. doi: 10.1002/path.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, Conejo-Garcia JR, Katsaros D, Gimotty PA, Massobrio M, Regnani G, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med. 2003;348:203–213. doi: 10.1056/NEJMoa020177. [DOI] [PubMed] [Google Scholar]
- 17.Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med. 2004;10:942–949. doi: 10.1038/nm1093. [DOI] [PubMed] [Google Scholar]
- 18.Yang I, Tihan T, Han SJ, Wrensch MR, Wiencke J, Sughrue ME, et al. CD8+ T-cell infiltrate in newly diagnosed glioblastoma is associated with long-term survival. J Clin Neurosci. 2010;17:1381–1385. doi: 10.1016/j.jocn.2010.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Safdari H, Hochberg FH, Richardson EP., Jr Prognostic value of round cell (lymphocyte) infiltration in malignant gliomas. Surg Neurol. 1985;23:221–226. doi: 10.1016/0090-3019(85)90086-2. [DOI] [PubMed] [Google Scholar]
- 20.Rossi ML, Jones NR, Candy E, Nicoll JA, Compton JS, Hughes JT, et al. The mononuclear cell infiltrate compared with survival in high-grade astrocytomas. Acta Neuropathol. 1989;78:189–193. doi: 10.1007/BF00688208. [DOI] [PubMed] [Google Scholar]
- 21.Heimberger AB, Abou-Ghazal M, Reina-Ortiz C, Yang DS, Sun W, Qiao W, et al. Incidence and prognostic impact of FoxP3+ regulatory T cells in human gliomas. Clin Cancer Res. 2008;14:5166–5172. doi: 10.1158/1078-0432.CCR-08-0320. [DOI] [PubMed] [Google Scholar]
- 22.Parsa AT, Waldron JS, Panner A, Crane CA, Parney IF, Barry JJ, et al. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat Med. 2007;13:84–88. doi: 10.1038/nm1517. [DOI] [PubMed] [Google Scholar]
- 23.Fecci PE, Ochiai H, Mitchell DA, Grossi PM, Sweeney AE, Archer GE, et al. Systemic CTLA-4 blockade ameliorates glioma-induced changes to the CD4+ T cell compartment without affecting regulatory T-cell function. Clin Cancer Res. 2007;13:2158–2167. doi: 10.1158/1078-0432.CCR-06-2070. [DOI] [PubMed] [Google Scholar]
- 24.Fecci PE, Mitchell DA, Whitesides JF, Xie W, Friedman AH, Archer GE, et al. Increased regulatory T-cell fraction amidst a diminished CD4 compartment explains cellular immune defects in patients with malignant glioma. Cancer Res. 2006;66:3294–3302. doi: 10.1158/0008-5472.CAN-05-3773. [DOI] [PubMed] [Google Scholar]
- 25.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prins RM, Soto H, Konkankit V, Odesa SK, Eskin A, Yong WH, et al. Gene expression profile correlates with T-cell infiltration and relative survival in glioblastoma patients vaccinated with dendritic cell immunotherapy. Clin Cancer Res. 2011;17:1603–1615. doi: 10.1158/1078-0432.CCR-10-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cooper LA, Gutman DA, Chisolm C, Appin C, Kong J, Rong Y, et al. The tumor microenvironment strongly impacts master transcriptional regulators and gene expression class of glioblastoma. Am J Pathol. 2012;180:2108–2119. doi: 10.1016/j.ajpath.2012.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reis RM, Konu-Lebleblicioglu D, Lopes JM, Kleihues P, Ohgaki H. Genetic profile of gliosarcomas. Am J Pathol. 2000;156:425–432. doi: 10.1016/S0002-9440(10)64746-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer-Puttlitz B, Hayashi Y, Waha A, Rollbrocker B, Bostrom J, Wiestler OD, et al. Molecular genetic analysis of giant cell glioblastomas. Am J Pathol. 1997;151:853–157. [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe K, Peraud A, Gratas C, Wakai S, Kleihues P, Ohgaki H. p53 and PTEN gene mutations in gemistocytic astrocytomas. Acta Neuropathol. 1998;95:559–564. doi: 10.1007/s004010050840. [DOI] [PubMed] [Google Scholar]
- 35.Burger PC, Pearl DK, Aldape K, Yates AJ, Scheithauer BW, Passe SM, et al. Small cell architecture--a histological equivalent of EGFR amplification in glioblastoma multiforme? J Neuropathol Exp Neurol. 2001;60:1099–1104. doi: 10.1093/jnen/60.11.1099. [DOI] [PubMed] [Google Scholar]
- 36.Pollack BP, Sapkota B, Cartee TV. Epidermal growth factor receptor inhibition augments the expression of MHC class I and II genes. Clin Cancer Res. 2011;17:4400–4413. doi: 10.1158/1078-0432.CCR-10-3283. [DOI] [PubMed] [Google Scholar]