Abstract
Pan-genome ortholog clustering tool (PanOCT) is a tool for pan-genomic analysis of closely related prokaryotic species or strains. PanOCT uses conserved gene neighborhood information to separate recently diverged paralogs into orthologous clusters where homology-only clustering methods cannot. The results from PanOCT and three commonly used graph-based ortholog-finding programs were compared using a set of four publicly available strains of the same bacterial species. All four methods agreed on ∼70% of the clusters and ∼86% of the proteins. The clusters that did not agree were inspected for evidence of correctness resulting in 85 high-confidence manually curated clusters that were used to compare all four methods.
INTRODUCTION
Next-generation sequencing allows large sets of bacterial genomes from the same species to be generated for multiple strain comparisons. The observation that for some species strains can acquire and lose large portions of their protein repertoire led to the concept of the pan-genome (1,2). The most fundamental pan-genome analysis is to compare differences in protein content between strains. In order to determine these differences, a correspondence between equivalent proteins in different strains must be established. The most common meaning of equivalent protein is a protein’s ortholog. Orthologs are defined as homologous genes that are related through speciation from a single ancestral gene, not through gene duplication (3,4). Orthologs tend to serve the same role and have the same function, particularly the more closely related the organisms are. Furthermore, for pan-genome analysis of closely related strains, ‘operational’, not functional, equivalence is more desirable than functional equivalence alone since, for example, two copies of a nearly identical protein are likely functionally equivalent, but could be under differential regulation. The copies under similar transcriptional regulation (i.e. in similar genomic neighborhoods) are likely to be the ones with similar operational equivalence; therefore, pan-genome analysis software should consider the genomic neighborhood of orthologous genes. When a gene is duplicated after speciation, or in species pan-genomes after strain differentiation, both copies of the gene are defined to be co-orthologs to the unduplicated gene in the other species or strains. For pan-genome analysis, we believe it is preferable to cluster only the co-orthologs with the same genomic context, but additional information should be reported indicating the co-ortholog relationship.
In general, determining orthologs is a hard problem (4–6) and has most often been investigated across species where evolutionary time has allowed for a great deal of protein sequence and genome context divergence. For greatly diverged species, genome context has been found to have little benefit for ortholog clustering (7). The key issue is distinguishing paralogs, homologous genes arising from gene duplications, from orthologs. Often, after gene duplication, paralogs diverge to take on different roles and functions. For diverged species, tree-based methods tend to perform best at ortholog clustering, albeit at the cost of being much less computationally efficient. The reason for this is that tree-based methods build multiple sequence alignments that can distinguish which amino acid residues are conserved within orthologs, but not between paralogs, even when the average pairwise alignment scores for orthologs versus paralogs may be indistinguishable. Graph-based methods, which rely on only the pairwise alignment scores, which are much more computationally efficient to generate, can suffer by comparison. For strains of the same species, the orthologous proteins tend to have little divergence and retain a conserved genome context. Paralogs that have diverged are easily distinguishable from the highly conserved orthologs by simple pairwise distances. Very recently duplicated paralogs are often indistinguishable even using tree-based methods, but are separable based on genome context. Pan-genome ortholog clustering tool (PanOCT) was designed to make use of this genome context or conserved gene neighborhood (CGN) information to better separate very recent paralogs.
There are a number of commonly used programs for determining orthologous gene clusters, but they were designed for clustering genes from distantly related eukaryotes, not closely related strains/species. These ortholog-finding programs consist of three conceptual methods: tree-based, graph-based and hybrid methods (4). Tree-based methods infer orthologs and paralogs by comparison of trees made with homologous genes to species trees. Graph-based methods use pairwise alignments to determine homology/distance between proteins to weight edges of the graph. Hybrid methods use a combination of tree- and graph-based methods. Mainly for computational efficiency, but also for availability, the graph-based InParanoid (8), OrthoMCL (9) and Sybil (10) ortholog clustering programs are often used for comparative genomic analysis (11–16).
PanOCT is a graph-based method, but differs from existing methods in its use of both the Basic Local Alignment Search Tool (BLAST) score ratio (BSR) (17) and CGN in a weighted scoring scheme to generate clusters containing single orthologous genes from each of multiple genomes and by detecting and accounting for potential frame-shifts. The concept of using the context of neighboring genes, that are themselves orthologous, to identify orthologs is not new (7,18); however, coupling CGN together with pairwise sequence identity and frame-shift detection to cluster orthologs in a single open-source application is novel. Algorithms have been developed that use both reciprocal best hit (RBH) and CGN, but either are used only as the back-end of a static database (ATGC, (19)), are used to score and visualize the genomic context of homology ‘pillars’ in a web browser (YGOB, (20)), or are functioning to re-cluster pre-computed ortholog/paralog clusters using CGN (IONS, (21)). Direct comparison with ATGC was not possible since the application was unavailable. PanOCT was compared with three popular graph-based programs: InParanoid (8), OrthoMCL (9) and Sybil (10) alone and in combination with IONS (21). GOB, the back-end CGN-detection script of YGOB (20), was obtained from the author. Using only ortholog clusters that were the same for InParanoid, OrthoMCL, Sybil and PanOCT as the pillars to input to GOB, the output of GOB was also compared with PanOCT.
InParanoid (8) tries to distinguish out-paralogs (i.e. duplications occurring before a species split) from in-paralogs (i.e. recent duplications after a species split) using a combination of RBH, also known as bi-directional best hit, and a heuristic clustering method for resolving overlapping groups of paralogs. A pairwise BLASTP cutoff score of 50 bits and an overlap cutoff of 50% are required for further consideration of orthology.
OrthoMCL (9) tries to distinguish in-paralogs from out-paralogs similarly to InParanoid. This program also uses RBH BLASTP matches to identify orthology, but uses a BLASTP P-value cutoff of 1 × 10−5 instead of the bit score cutoff and does not consider the length of the match. Potential orthologous and paralogous protein relationships are converted into a graph with weighted edges. The resulting graph is used as input to the Markov Cluster algorithm (22) to attempt to separate orthologs from paralogs.
Sybil (10) clusters are computed in a two-step process: Jaccard coefficient-based clustering of the proteins within a genome to determine paralogs and RBH BLASTP match clustering of the resulting Jaccard clusters (JAC) between genomes to determine orthologs. The Jaccard clustering step computes a similarity coefficient from filtered intra-genome unidirectional pairwise BLASTP matches (E-value of at most 1 × 10−5 and a percent identity of at least 80%), resulting in clustering of in-paralogs called JACs. RBH matches of JACs from different genomes are then clustered to form Jaccard orthologous clusters. Similar to InParanoid and OrthoMCL, Sybil clusters in-paralogs with orthologs, but the JAC parameters can be set to effectively exclude Jaccard clustering results, creating ortholog-only clusters based solely on the RBH BLASTP matches.
PanOCT uses BLASTP matches and CGN to predict orthologous clusters for pan-genomes. CGN is defined as the conservation of gene order and orientation within the genomes of closely related species. PanOCT is specifically designed for pan-genome analysis of closely related species/strains where CGN can be effectively used to distinguish groups of paralogs into separate clusters of orthologs (7); however, it will also work on analysis of more distantly related microbial species, but CGN will be of less benefit.
MATERIALS AND METHODS
System and software requirements
PanOCT was written in PERL (http://www.perl.org (1 August 2012, date last accessed)) and tested using PERL version 5.10 on Linux CentOS and Mac OS X 10.6 operating systems. To perform BLAST searches, either National Center for Biotechnology Information (NCBI) BLASTALL (23) version 2.2.10 or later (ftp://ftp.ncbi.nih.gov/blast/executables/release/ (1 August 2012, date last accessed)) or WUBLAST 2.0 (now called AB-BLAST available at http://blast.advbiocomp.com (1 August 2012, date last accessed)) are required.
Input requirements
PanOCT requires four input files. The first is either a NCBI (options -m 8 or 9) or WUBLASTP (using the btab program) tabular output file consisting of all-versus-all BLASTP searches of all predicted proteins in each genome to be analyzed. The second input requirement is a text file containing unique genome identifiers, one identifier per line, to determine which genome is to be treated as the reference genome in the output files and which genomes to include in the analysis. The genome identifier can be associated with specific proteins in two ways: (i) by placing the genome identifier after the protein identifier (e.g. NT08AB0001-GENOME_IDENTIFIER) or (ii) in the gene attribute file. The gene attribute file is the third required input file. It is a tab-delimited file containing the following data: contig id, protein identifier (e.g. locus), 5′-coordinate, 3′-coordinate, annotation and genome identifier. The final input file requirement is the protein fasta file used in the all-versus-all BLASTP searches. The protein fasta file is used by PanOCT to calculate the length of each protein, which is necessary in order to compute the BSR.
\In addition to the input file requirements mentioned above, PanOCT has a number of configurable command-line options: amino acid percent identity cutoff (default is 35%), BLAST E-value cutoff (default is 10−5), minimum percent match length of subject and query (default is 1%), frame-shift overlap parameter (default is 1.33; can be disabled), the number of amino acids at the beginning or end of a match that can be missing and still be considered a full length match between 0 and 100 (default is 20) and the number of blast matches needed to confirm a protein fragment/frame-shift (default is 1).
Output
PanOCT produces seven tab-delimited text output files plus a runtime report file. The most informative output files are the match_table and match_table_id files, which contain the ortholog clusters, one cluster per row. Each column contains protein identifiers with the first column beginning with the reference genome, followed by subsequent genomes ordered as instructed in the genome identifier file. The match_table_id file lists the percent identity for each protein to the reference in addition to the protein identifiers. The third output file has the following fields: locus identifier, annotation and each subsequent column containing the BSR (17). The fourth file is the frame-shift report, showing for each protein fragment the identity of retained and ignored fragments that are considered part of the same frame-shifted gene/pseudogene. Additional optional output files are available and are documented in the README.txt packaged with the distributed tarball. A detailed description of each output file is included with the source code.
PanOCT overview
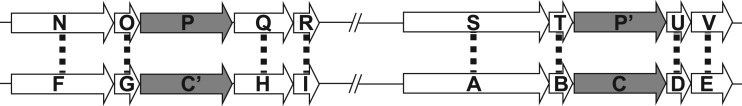
All ortholog clustering methods start with a measure of protein similarity/homology based on alignment scoring between the proteins. The selection of orthologs is clear when only one protein per genome is homologous to each other (although even in this case compensating gene losses in each organism could cause non-orthologs (i.e. out-paralogs) to be considered orthologous). However, the choice is more complicated when multiple proteins per genome are homologous. When the species are closely related, CGN can be used as a contextual clue to discriminate between orthologs and paralogs. For example, two proteins from the same genome, P and P′, are homologs and have homology to proteins C and C′ from another genome. Based on homology alone, if P is more similar to C than to C′, and C is more similar to P than to P′, C might be assigned the ortholog of P based on RBH. However, by using CGN to distinguish orthologs when the proteins are nearly identical in addition to RBH, C′ can be assigned as the ortholog of P and C the ortholog of P′ (Figure 1).
Figure 1.
An example of how CGN can be used to cluster paralogous genes into orthologous pairs. Open arrows indicate proteins, with dotted lines illustrating best BLAST matches. Gray arrows indicate paralogous proteins with multiple high-identity BLAST matches. The genes on top are from one genome and those on the bottom from a second genome. The slashes indicated that there are genes in between not drawn.
There is circularity in the above example where CGN evidence for a choice of ortholog is based on prior identification of the orthologs for the neighboring genes. To address this problem, a measure of likely orthology using only homology information must be generated before determining the final orthology using CGN information. This measure of orthology is then combined within a window surrounding each potential ortholog pair (POP) to assign a weighted score (WS), including both homology and CGN measures.
Flowchart
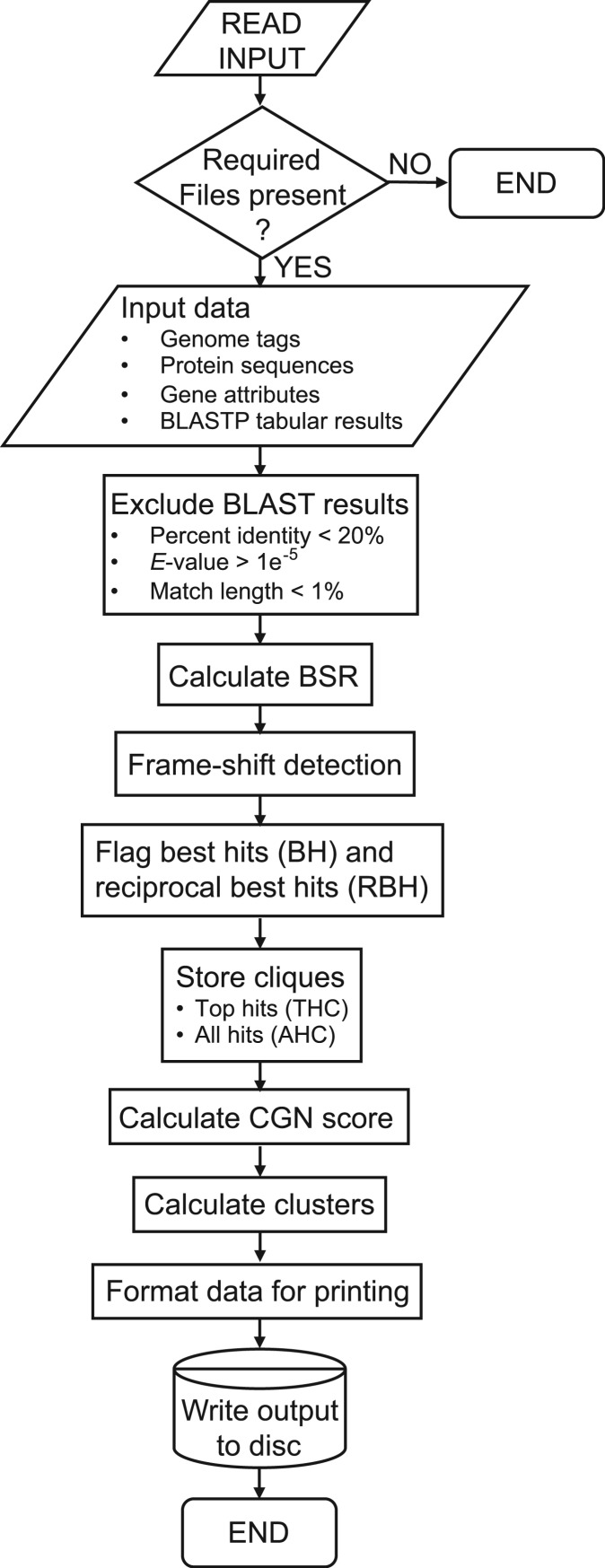
PanOCT initiates by reading in the four input files described above (Figure 2). The set of pairwise BLAST matches that pass defined minimal cutoff criteria are considered valid BLAST matches (assigned nonzero homology) and become directed edges (from the query protein to the target protein) in the homology graph. Nodes in the graph represent the proteins. Edges represent directed homology relationships from a query protein node to a target protein node. Edges between nodes in the same genome are used for some of the homology scoring criteria, but are not used for ortholog clustering in order to exclude paralogs. The BSR is computed for each valid BLAST alignment and stored for each edge. The BSR is the bit score of the BLAST alignment divided by the bit score of the query protein aligned to itself. Potential frame-shifted genes are then identified (see ‘Frame-shift detection’ section). The outgoing edges from each node are sorted by the BSR values to determine and flag the best hit (BH) edge for the query protein to a target protein in each genome. A RBH is found when there exists a pair of oppositely directed BH-directed edges between a pair of nodes. Both oppositely directed BH edges are also flagged as RBH edges. The top RBH edges for a node are defined to be the set of outgoing RBH edges that have higher BSR values than any non-RBH outgoing edge from that node including edges to the same genome. The top RBH edges are then tested to see if they form a clique (a completely connected subgraph, where undirected edges exist between every node in the subgraph). The edges within the clique are treated as undirected since each node within the clique has two oppositely directed edges due to being RBH. Cliques are defined as a Top Hits Clique (THC) if a set of nodes and their top RBH edges form a clique of size greater than two and further defined as an All Hits Clique (AHC) if the top RBH edges are also the only edges between all nodes in the clique. Next, a homology score (HS) is computed and used in the calculation of a CGN score (see ‘CGN score’ section and Figure 3A). The CGN score is assigned for each directed edge in the graph between POPs (Figure 3B). Edges are filtered out prior to clustering that are not RBH by CGN score to remove ‘noise’ edges. Ortholog clusters are computed by hierarchically agglomerating protein nodes greedily using the highest scoring CGN edges first. Clusters grow as the next largest CGN score edge connects the clusters containing the query and target proteins for that edge. This merging of clusters is not allowed if the resulting cluster would have more than one protein from the same genome. The final step is to format the data for writing to any of the optional output files.
Figure 2.
Flow chart of PanOCT.pl logic. Standard symbols for constructing flow charts were used.
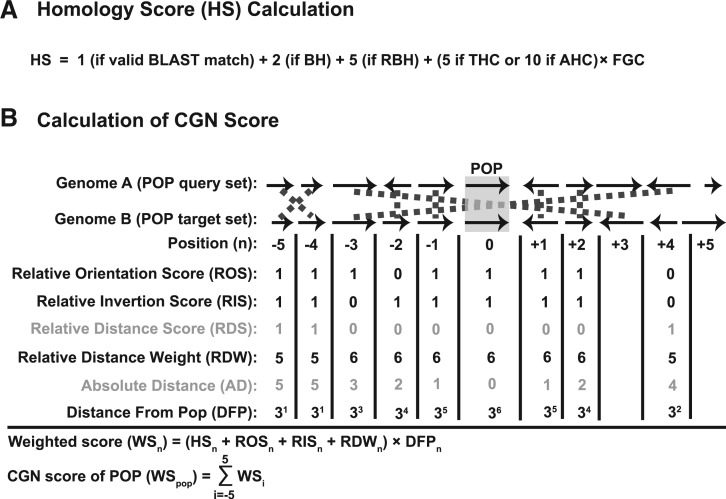
Figure 3.
Calculation of the HS (A) and an example of how the CGN is computed (B).
Frame-shift detection
Even for the four ‘complete’ genomes extensively analyzed for this paper, a protein from one genome would often match adjacent protein fragments in a different genome due to frame-shift sequencing/assembly errors or the presence of pseudogenes. To identify potential frame-shifted genes, PanOCT looks for BLAST matches of one protein to more than one adjacent protein in the appropriate order, orientation and with a minimal overlap. To distinguish between protein fragments and tandem protein duplication, PanOCT tests if the amount of the target protein covered by non-overlapping sequence is significantly more than that covered by overlapping sequence. The ratio of overlapping to non-overlapping coverage is a user-definable parameter. In situations when sequencing or assembly error may have resulted in the fusion of neighboring genes, a voting scheme is used to determine if adjacent protein fragments should be combined or left as separate proteins. PanOCT will retain the protein fragment with the longest BLASTP match length to another protein for ortholog clustering while ignoring those fragments with shorter BLASTP match lengths.
Homology score
The HS is set to one for a valid BLAST match (Figure 3A). The HS is incremented by two if the target protein is the BH for the query protein for the target protein’s genome. The weight is further incremented by five if the edge is a RBH. A RBH is when the target protein is a best BLAST match for the query protein and reciprocally the query protein is a best BLAST match for the target protein as sorted by BSR. The top RBH edges are defined for a node to be all outgoing directed RBH edges, which when sorted by BSR come before any outgoing non-RBH edges, including paralog edges. By definition, paralog edges cannot be RBH edges since RBH edges can only be between proteins in different genomes. Each query protein will have at most one directed RBH edge per target genome. There will be a symmetric oppositely directed RBH edge from the target protein to the query protein. We treat this pair of directed RBH edges as an undirected edge for clique determination. If a set of nodes and their top RBH edges form a THC of size greater than two (i.e. more than pairwise), the HS is incremented by five times the fraction of genomes in the clique (FGC; Figure 3A). However, if the top edges are also the only edges (an AHC), the HS is incremented by 10 times the FGC.
CGN score
For each possible ortholog edge and the POP it connects in the graph, we compute a CGN score (Figure 3B). The CGN score for a POP is a weighted combination of the HSs for a fixed number N (currently set to 5) of genes upstream and downstream of both the query and target proteins in a POP. These weights are based on: relative orientation score (ROS), relative inversion score (RIS), relative distance weight (RDW) and distance from the POP (DFP, Figure 3B). For each protein in the POP query set, every homology edge to the POP target set is evaluated. The POPs are oriented in the same direction for calculating relative orientation and relative inversion scores.
For each homology edge, the WS, which is initialized to be equal to the HS, is incremented by one (via the ROS) if the protein from the query set has the same relative orientation as the protein from the target set. If the query set protein is on the same strand as the POP query protein, then the target set protein should be on the same strand as the POP target protein. The WS is further incremented by one if the query set protein and the target set protein both have not been inverted relative to the POP (measured by the RIS). The RIS is set to one if both the query and target set proteins are upstream of the POP. Likewise, the RIS has a value of one if both the query and target set proteins are downstream of the POP.
In a well-conserved gene neighborhood, orthologs are expected to be in exactly the same relative DFP, unless broken by insertion, deletion or inversion events. A relative distance score (RDS) is assigned to penalize query/target set proteins that are not the same number of genes from the POP. The relative distance for a query or target set protein is the number of proteins it is away from the POP, defined as the position number (n; Figure 3B). The RDS is computed as the absolute value of the difference between the positions of the query and target set proteins. Using this penalty, RDS is converted to a RDW (N + 1 minus the RDS; Figure 3B). By definition, the match between the POP query protein and the POP target protein will receive maximal increments from the ROS, RIS and RDW.
Since the CGN score is used as an attempt to break near ties in the HSs, the pairwise WS for each query set protein should not be equally weighted but rather focused on the POP query protein itself and its nearest neighbors. To achieve this, the weight of the WS is exponentially decreased the further away the query/target set proteins are from the POP via the DFP weight (Figure 3B). Each query set protein may have more than one pairwise WS if paralogs exist within the target set proteins. To avoid double counting either query or target set paralogs, the larger of the two WSs is used. The CGN score of the POP is computed as the sum of the pairwise WSs for the entire set of query proteins.
Validation
The complete genome sequences of four Acinetobacter baumannii strains were downloaded from GenBank: ATCC 17978 (CP000521-523), AYE (CU459137-141), SDF (CU468230-233) and AB0057 (CP001182-1183). To obtain consistent structural annotation predictions across genomes, each genome was run through the JCVI annotation pipeline (24,25). These gene predictions were used to generate files containing the combined protein sequences of all four genomes and gene attributes for PanOCT (Supplementary Figures S1 and S2, respectively). NCBI BLASTP results were generated (E-value cutoff 1 × 10−5, filter off) and used by each of the four clustering methods. Each clustering method was run using default parameters with the exception of Sybil, where the Jaccard-clustering P-value was set to –1 to eliminate the inclusion of paralogs, which reduces Sybil to a simple RBH algorithm. An increase in the inflation parameter of OrthoMCL failed to exclude paralogs, so the default of 1.0 was maintained (data not shown). Cluster results from each of the clustering methods were compared. Clusters where all four methods agreed were assumed to be largely correct and uninformative for comparing relative performance. All other clusters were examined using ClustalW multiple sequence alignments, neighbor-joining trees, high-quality functional annotation evidence (HMM matches to equivalog level PFAM/TIGRFAM families (17)) and genomic context as evidence of orthology. This resulted in only 85 manually curated clusters containing 328 proteins that we felt were clustered with high confidence. The use of genomic context (CGN) as supporting evidence certainly biases the results toward PanOCT, which is the only method of the primary four to make use of CGN. That is why IONs and GOB, which also use CGN, were included in comparisons. These 85 manually curated clusters became the reference set used to evaluate the performance of each clustering method.
The clusters from each of the four primary ortholog clustering methods were given as input to IONs for four different runs. The set of clusters where all four primary methods agreed were used as the high confidence clusters or ‘pillars’ to be input to GOB.
The performance of each orthology detection method was evaluated using sensitivity (TP/(TP + FN)) and the positive predictive value (PPV; TP/(TP + FP)), where TP is the number of true positives, FN is the number of false negatives and FP is the number of false positives. The TP, FN and FP values were calculated over the set of pairwise orthology assertions inherent in each reference cluster. For example, if a reference cluster contained four orthologs (A, B, C and D), there would be six pairwise orthology assertions (A-B, A-C, A-D, B-C, B-D and C-D). For a clustering method, a TP is when the clustering method’s clusters inherently assert a pairwise orthology which is in the reference set, a FP is when the clustering method’s clusters inherently assert a pairwise orthology that is not in the reference set but at least one of the two proteins in the pairwise orthology is present in the reference set, and a FN is when the clustering method’s clusters do not inherently assert a pairwise orthology, which is present in the reference set (Supplementary Figure S3). Clustering methods were given credit for including a protein in a cluster when they included at least one protein fragment from that protein for frame-shifted genes/pseudogenes. Clusters were not penalized if they included multiple protein fragments from the same frame-shifted gene/pseudogene.
To determine the hardware demands of PanOCT compared with the other methods used in this study, resource statistics for each clustering method were tracked by submitting independent jobs to an Oracle Grid Engine (Oracle Corporation, Redwood Shores, CA, USA) grid, using the qacct command to gather information for each run. A total of 25 runs per method were launched. The input for the first runs contained three Escherichia coli genomes, whereas the final runs contained 25 genomes. Each run contained genomes used in the previous run plus one additional genome. All four methods were run on the same order of genomes to eliminate artifacts due to order of addition.
RESULTS
Protein clusters were computed for four clustering methods [PanOCT (Supplementary Figure S4), OrthoMCL (Supplementary Figure S5), InParanoid (Supplementary Figure S6) and Sybil (Supplementary Figure S7)] for all proteins in four A. baumannii genomes, and results of the differences were evaluated using automated measures and manual inspection. Each of the four methods differed in the number and size of clusters they produce (data not shown). All four methods produced similar numbers of clusters containing between one and four loci with PanOCT and Sybil containing more singleton clusters than either InParanoid or OrthoMCL (data not shown). Only OrthoMCL and InParanoid produced clusters containing more than four members, with the largest OrthoMCL cluster containing 213 members (Table 1). Every cluster with greater than four loci contained paralogs, whereas only 1–8% of clusters with one to four loci contained paralogs. OrthoMCL produced some clusters with no orthologs, only paralogs; whereas, InParanoid will only add paralogs if orthologs are also present.
Table 1.
Comparison of clusters containing paralogs
| No. of members | InParanoid |
OrthoMCL |
||
|---|---|---|---|---|
| Clusters | Paralogs n (%) | Clusters | Paralogs n (%) | |
| 1 | 1961 | 1415 | ||
| 2 | 523 | 527 | 42 (8) | |
| 3 | 849 | 12 (1) | 797 | 25 (3) |
| 4 | 2240 | 22 (1) | 2310 | 33 (1) |
| 5 | 40 | 40 (100) | 62 | 62 (100) |
| 6 | 17 | 17 (100) | 20 | 20 (100) |
| 7 | 10 | 10 (100) | 11 | 11 (100) |
| 8 | 4 | 4 (100) | 5 | 5 (100) |
| 9 | 2 | 2 (100) | 1 | 1 (100) |
| 10 | 2 | 2 (100) | ||
| 11 | 1 | 1 (100) | 3 | 3 (100) |
| 15 | 2 | 2 (100) | ||
| 16 | 1 | 1 (100) | ||
| 17 | 1 | 1 (100) | ||
| 18 | 1 | 1 (100) | 1 | 1 (100) |
| 22 | 1 | 1 (100) | ||
| 23 | 1 | 1 (100) | 1 | 1 (100) |
| 26 | 1 | 1 (100) | ||
| 29 | 1 | 1 (100) | ||
| 144 | 1 | 1 (100) | 1 | 1 (100) |
| 213 | 1 | 1 (100) | ||
| Total | 5653 | 114 (2) | 5161 | 212 (4) |
Comparing cluster membership
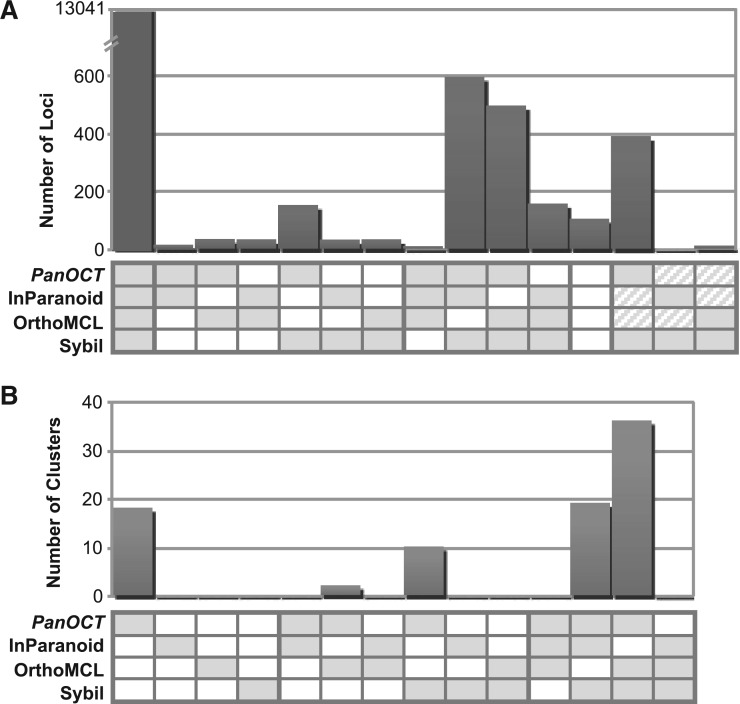
Except when all four methods agree, it is hard to directly compare clusters. This is because members of a single cluster from one clustering method could correspond to multiple clusters from another method, which may in turn correspond to different clusters from the original method. Therefore, instead of comparing clusters to evaluate the results of each clustering method, the cluster membership for each protein was evaluated. For each protein, two methods agreed if the protein was included in clusters with identical membership and disagreed otherwise. Of 6710 total non-redundant clusters containing 15 180 proteins, all four methods agreed for 86% of proteins (13 041) in 69% of the clusters (4631; Figure 4A). Three methods agreed and one disagreed: PanOCT, InParanoid and Sybil agreed for 4% of proteins; PanOCT, OrthoMCL and Sybil agreed for 3%; InParanoid, OrthoMCL and Sybil agreed for 1% and PanOCT, InParanoid and OrthoMCL agreed for <1% of proteins (Figure 4A).
Figure 4.
Agreement/disagreement between how proteins are clustered by the four methods for the entire set of clusters (A) and for the 85 manually curated clusters (B). The number of proteins (A) or clusters (B) that are in agreement for each possible subset of the four methods is graphed. Each subset pattern is indicated with shaded boxes for agreement and open boxes for disagreement. For example, when there are two shaded boxes and two open boxes the two shaded methods agree and the two open methods disagree with all three other methods; diagonal lines in a box indicate that while the two methods with diagonal lines disagree with the two shaded methods they agree with each other.
Comparing to reference clusters
To directly compare and evaluate the behavior of each method, a reference set of 85 manually curated clusters was generated out of the set of clusters shown to disagree among the methods. Clustering method results were compared back with the reference set and cluster agreement or disagreement was scored (Figure 4B). Of the 85 reference clusters, PanOCT agreed with 85 (100%), InParanoid agreed with 19 (22%), OrthoMCL agreed with 38 (45%) and Sybil agreed with 65 (76%). These results can be split into 15 (24–1) possible combined cluster prediction patterns of the four methods (Figure 4B). PanOCT, OrthoMCL and Sybil made the same cluster prediction for the largest number (36 or 42%) of clusters in agreement with the reference set. The next three major patterns of agreement with the reference clusters were PanOCT, InParanoid and Sybil (19 or 22%), PanOCT alone (18 or 21%), and finally PanOCT and Sybil (10 or 12%) (Figure 4B).
Performance
The performance of each method was further compared with the 85 reference clusters, using both sensitivity and PPV. PanOCT had the highest PPV of 1.000, while Sybil, InParanoid and OrthoMCL had PPVs of 0.958, 0.766 and 0.699, respectively (Table 2). PanOCT also had the greatest sensitivity (1.000), followed by OrthoMCL with 0.922, then Sybil and InParanoid at 0.863 and 0.692, respectively. Consistent with the findings in Table 1 where OrthoMCL had the largest number of paralogs in clusters, OrthoMCL had the highest FP rate of all four methods. InParanoid was the worst performer against the reference clusters, having the lowest number of TP, and the second highest FP rate. It is a little misleading to report sensitivity and PPV for only the 85 reference clusters, representing clusters that differed among the four clustering methods, since this is only a fraction of the number of clusters where all four methods agreed. If we presume that the clusters where all four methods agreed are overwhelming correct; hence, treating them as true positives, the PPV values for an expanded set of 4716 clusters (85 + 4631) were 1.000, 0.999, 0.995 and 0.990 for PanOCT, Sybil, InParanoid and OrthoMCL, respectively, and the sensitivity values were 1.000, 0.997, 0.993 and 0.998 for PanOCT, Sybil, InParanoid and OrthoMCL, respectively (Table 2).
Table 2.
Evaluating method performance using PPV and sensitivity measurements
| Method | TP | FP | FN | PPV | Sensitivity |
|---|---|---|---|---|---|
| Four clustering methods againt 85 reference clusters | |||||
| PanOCT | 401 | 0 | 0 | 1.000 | 1.000 |
| Sybil | 345 | 15 | 55 | 0.958 | 0.863 |
| InParanoid | 269 | 82 | 120 | 0.766 | 0.692 |
| OrthoMCL | 365 | 157 | 31 | 0.699 | 0.922 |
| Four clustering methods against expanded reference clusters | |||||
| PanOCT | 16 074 | 0 | 0 | 1.000 | 1.000 |
| Sybil | 16 018 | 15 | 55 | 0.999 | 0.997 |
| InParanoid | 15 942 | 82 | 120 | 0.995 | 0.993 |
| OrthoMCL | 16 038 | 157 | 31 | 0.990 | 0.998 |
| Four Clustering methods plus IONS or GOBS against 85 reference clusters | |||||
| PanOCT IONS | 398 | 0 | 3 | 1 | 0.99 |
| Sybil IONS | 342 | 6 | 60 | 0.98 | 0.85 |
| InParanoid IONS | 266 | 52 | 123 | 0.84 | 0.68 |
| OrthoMCL IONS | 363 | 49 | 33 | 0.88 | 0.92 |
| GOBS | 325 | 1 | 62 | 1.00 | 0.84 |
CGN post-processing with IONS and GOB
To independently confirm that CGN improves clustering of orthologs, the previously determined clusters from each clustering method and the consensus clusters where all four methods agree were used as input for IONS (21) and GOB (20), respectively. IONS was able to correct some initial clusters for OrthoMCL (Supplementary Figure S8) and InParanoid (Supplementary Figure S9) by splitting clusters with paralogs into ortholog-only clusters using CGN (Table 2). IONs seemed to be particularly tuned for OrthoMCL clusters. For Sybil (Supplementary Figure S10) and PanOCT (Supplementary Figure S11), IONS could not improve clustering since there were no paralogs in the clusters but did split a few good clusters, degrading the performance slightly. IONS did not achieve the same performance as PanOCT when starting with clusters from another method (Table 2). GOB was able to use the confident ortholog clusters where all four primary methods agreed, to better determine separation of paralogs using CGN (Supplementary Figure S12), but still did not match PanOCT’s performance, measured by Sensitivity (Table 2).
Evaluation of differences
To better understand the reason for the observed differences in performance, each prediction pattern with disagreement to the reference clusters was manually evaluated and summarized in Supplementary Table S1. About 50% of the 85 references clusters were incorrectly identified due to the presence of paralogs, while the remaining 50% of reference clusters were missed because of differences in protein length (e.g. different start sites, truncations and splitting due to frame-shifts). There were several examples where OrthoMCL (Figure 5A), InParanoid (Figure 5B) or both failed to separate significantly diverged paralogs into ortholog clusters when Sybil and PanOCT do. It is not clear from our analysis why OrthoMCL differed from InParanoid on which diverged paralogs to include in a cluster. Where PanOCT differed in paralog separation from Sybil is for less diverged paralogs where homology is not sufficient and CGN information must be used (Figure 6). Looking outside of the 85 reference clusters, there were a few instances where PanOCT and/or Sybil seemed to make an arbitrary choice between paralogs when no convincing CGN or homology information existed (data not shown).
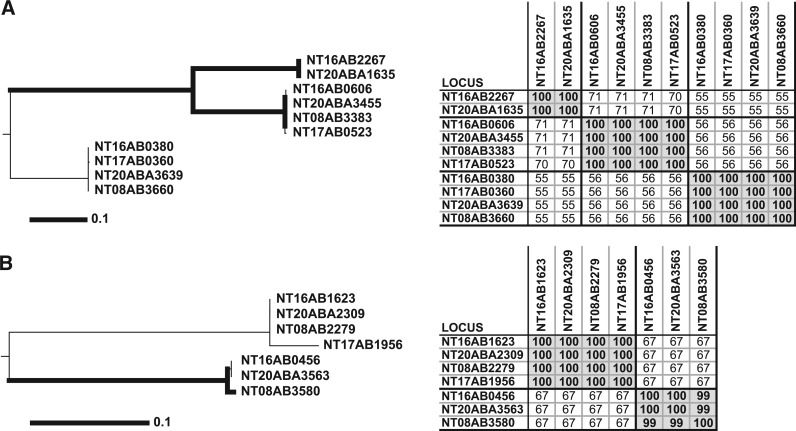
Figure 5.
Separation of divergent paralogs into orthologous clusters. The left panels denote consensus Neighbor-joining trees from 100 bootstrap replicates as previously described (26). The thick lines infer the strength of bootstrap values >74. The scale bar represents the number of amino acid substitutions. The panels on the right show tables of pairwise BLAST protein percent identities. OrthoMCL (A) and InParanoid (B) grouped every protein depicted into two large clusters (one in A and another in B), while the other three methods produced clusters identical with the phylogenetic trees (three in A and two in B). Protein percent identity was sufficient to group these proteins into unambiguous clusters (tables on right).
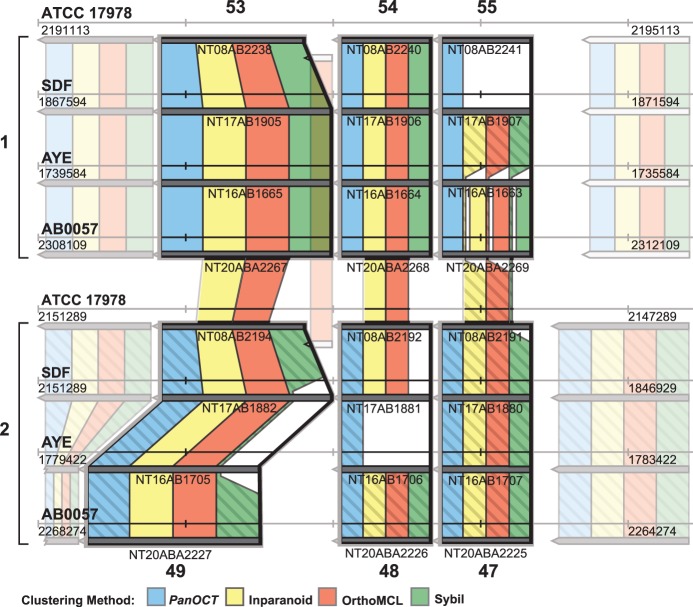
Figure 6.
Separation of paralogs into orthologous clusters using CGN. Cluster results of each of the four methods compared with six reference clusters. Reference clusters are outlined in bold (reference clusters 53, 54, 55, 49, 48 and 47 from Supplementary Table S1). Note that reference clusters 53, 54 and 55 are located in a chromosomal region different from reference clusters 49, 48 and 47. These regions are labeled 1 and 2, respectively, and method cluster results are colored either solid or hashed to help differentiate.
Scalability
Since PanOCT was created for pan-genome analysis, a much larger set of bacterial genomes was used to test the scalability of the PanOCT software. A graph of the reported maximum memory usage showed that PanOCT used more memory per genome than the other three methods (Supplementary Figure S13A). PanOCT memory usage is unchanged until the sixth genome is added, with a usage of ∼0.25 GB per genome, maxing out at 0.5 GB per genome by the 25th genome. As a result of an in-memory data storage strategy, PanOCT is able to finish orders of magnitude faster than Sybil and InParanoid, and in a fraction of the time it takes OrthoMCL, when run on identical data (Supplementary Figure S13B).
Robustness
To show that PanOCT is robust for large datasets, PanOCT was run on a set of 60 A. baumannii strains (Supplementary Table S2), including the original four used for analysis in this article. Clusters formed with the original four strains were compared with the clusters from 60 strains (Supplementary Figure S14). The clusters generated from the pan-genome analysis of 60 strains were projected onto the original four strains by removing all proteins not from the original four strains from the clusters and then compared with the original clustering results. A robust clustering method should minimize the clusters that change (split or merge) when new strains are added. For the 85 reference clusters, 1 cluster changed. For the 4361 clusters where all four methods agreed, 100 clusters changed. For the entire set of 5865 PanOCT clusters, 262 clusters changed. The small number of clusters that changed indicates that PanOCT is generally a robust clustering method. Almost all of the clusters that changed were joined by weak homology with a lack of CGN evidence, suggesting that in these instances, PanOCT may be too aggressive in assigning orthologs.
DISCUSSION
Pan-genome analysis is simplified when clear orthologs can be determined and separated from paralogs. PanOCT utilizes CGN information to separate recently diverged paralogs into orthology clusters where other, homology-only clustering methods cannot.
Systemic differences in clustering tendencies between the methods were analyzed by examining clusters where the methods differed. The tendency of OrthoMCL and InParanoid to agree is, in part, due to the inclusion of paralogous proteins in their clusters, while PanOCT and Sybil tend to agree because they do not. Notably, OrthoMCL and InParanoid often disagree on which paralogs to include when including paralogs (Figure 4A). OrthoMCL formed clusters with only paralogs from a single genome, while InParanoid only formed clusters containing paralogs if there was at least one ortholog from another genome. It is not clear when, if at all, paralogs should be included in clusters for pan-genome analysis. Sybil often failed to include closely related paralogs in clusters because their protein sequences were too similar to be separated by RBH alone, creating singleton clusters for the paralogs, justifying the use of CGN to aid in clustering.
Recruiting paralogs into clusters negatively impacted the PPV of OrthoMCL and InParanoid compared with Sybil and PanOCT, through an increase in the number of false positives. Therefore, a more direct comparison can be made by comparing PanOCT to Sybil (with Jaccard clustering turned off), which do not include paralogs. This then becomes a comparison of RBH plus CGN versus RBH alone. Both methods have excellent PPVs, but differ mainly in sensitivity. PanOCT had greater sensitivity than Sybil primarily because it was able to place proteins with very similar identities into clusters based on the conservation of gene order surrounding these loci. This provides support for the use of CGN in addition to RBH to cluster orthologs from closely related species.
One of our four test genomes contained a large number of transposons, which were frequently inserted into a gene creating two protein fragments. PanOCT included the fragment with the more conserved CGN in some cases where the other methods favored the longer fragment or in the case of OrthoMCL would include both fragments in the same cluster. InParanoid applies a protein length constraint so that a match is not considered if it is less than half the length of the longer protein. This means that InParanoid will not include as many protein fragments in clusters as the other three methods do. PanOCT was the most permissive in allowing short fragments (i.e. from truncation events), causing some differences compared with the other three methods. Future revisions of PanOCT could be made to treat protein fragments created by transposon insertions in the same way it handles frame-shifted/pseudogene protein fragments, by ignoring the shorter fragment for clustering purposes and outputting this information to a file.
Further investigation is needed to determine the appropriate orthology clustering when little or no CGN evidence is present. Some interesting occurrences were observed in the test genomes, which raise questions as to when orthology should be asserted and if in some cases clusters should be created when orthology is not asserted. There were several observed cases where a gene is duplicated at a point of genome rearrangement with strong CGN present on only one side of each of the duplicated proteins (data not shown). Should this duplicated gene be included in a cluster as an in-paralog since it cannot be separated using CGN? There were cases where strong CGN existed, but a protein in the middle of the CGN was more diverged in one genome than the others: is this a rapidly diverging ortholog or a horizontal transfer/homologous recombination? In other cases, a string of highly diverged proteins with reasonable CGN were observed, which may represent a horizontally transferred cassette. OrthoMCL tends to put protein fragments due to frame-shifts, pseudogenes or transposon insertions into the same cluster, whereas PanOCT recognizes and outputs these situations explicitly.
PanOCT currently creates ortholog clusters containing at most one protein from each genome. For highly similar proteins, which could in some cases be co-orthologs, PanOCT uses CGN to make a choice of which to include in a cluster. The intent is to cluster proteins that are most likely to be operationally equivalent. We plan to output information about these highly similar proteins in a file separate from the cluster output in a future release of PanOCT.
In conclusion, orthology detection programs designed for the purpose of comparing the protein content of distantly related eukaryotes, such as OrthoMCL and InParanoid, might not be well suited to bacterial pan-genome studies, particularly for studies including draft genome sequences. Although all four methods compared in this study agreed on ∼70% of the clusters and ∼86% of the proteins, PanOCT, by using CGN, outperformed three other clustering methods for a manually curated set of reference clusters. IONS and GOB were able to take some clustering information as input and use CGN to improve performance, but did not match PanOCT’s performance. Although we believe that PanOCT outperforms other ortholog clustering tools, there is still room for improvement, particularly in being more conservative at ortholog assertion to make the clustering more robust to the addition of more genomes.
AVAILABILITY
The PanOCT source code as well as the input and output files used in this study can be freely obtained at http://panoct.sourceforge.net/ (2 August 2012, date last accessed) under version 3 of the GNU General Public License (http://www.gnu.org/licenses/ (2 August 2012, date last accessed)).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2 and Supplementary Figures 1–14.
FUNDING
This project was funded in whole or part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under contract numbers N01-AI30071 and / or HHSN272200900007C. Funding for open access charge: HHSN272200900007C.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
Thanks go to Matthew LaPointe for help with graphic design and Claudia Haywood for legal assistance with the open source license. We also thank Mare-Line Seret for help with running IONS and Kevin Byrne for providing GOB.
REFERENCES
- 1.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl Acad. Sci. USA. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tettelin H, Riley D, Cattuto C, Medini D. Comparative genomics: the bacterial pan-genome. Curr. Opin. Microbiol. 2008;11:472–477. doi: 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 3.Fitch WM. Distinguishing homologous from analogous proteins. Syst. Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- 4.Kuzniar A, van Ham RC, Pongor S, Leunissen JA. The quest for orthologs: finding the corresponding gene across genomes. Trends Genet. 2008;24:539–551. doi: 10.1016/j.tig.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 5.Kristensen DM, Wolf YI, Mushegian AR, Koonin EV. Computational methods for Gene Orthology inference. Brief. Bioinform. 2011;12:379–391. doi: 10.1093/bib/bbr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gabaldon T. Large-scale assignment of orthology: back to phylogenetics? Genome. Biol. 2008;9:235. doi: 10.1186/gb-2008-9-10-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huynen MA, Bork P. Measuring genome evolution. Proc. Natl Acad. Sci. USA. 1998;95:5849–5856. doi: 10.1073/pnas.95.11.5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Remm M, Storm CE, Sonnhammer EL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 9.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome. Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crabtree J, Angiuoli SV, Wortman JR, White OR. In: Gene Function Analysis. Ochs MF, editor. Totowa, NY: Humana Press Inc; 2007. pp. 93–108. [Google Scholar]
- 11.Zhang A, Yang M, Hu P, Wu J, Chen B, Hua Y, Yu J, Chen H, Xiao J, Jin M. Comparative genomic analysis of Streptococcus suis reveals significant genomic diversity among different serotypes. BMC Genomics. 2011;12:523. doi: 10.1186/1471-2164-12-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sucgang R, Kuo A, Tian X, Salerno W, Parikh A, Feasley CL, Dalin E, Tu H, Huang E, Barry K, et al. Comparative genomics of the social amoebae Dictyostelium discoideum and Dictyostelium purpureum. Genome Biol. 2011;12:R20. doi: 10.1186/gb-2011-12-2-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazzaglia A, Studholme DJ, Taratufolo MC, Cai R, Almeida NF, Goodman T, Guttman DS, Vinatzer BA, Balestra GM. Pseudomonas syringae pv. actinidiae (PSA) isolates from recent bacterial canker of Kiwifruit outbreaks belong to the same genetic lineage. PLoS One. 2012;7:e36518. doi: 10.1371/journal.pone.0036518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biggs PJ, Fearnhead P, Hotter G, Mohan V, Collins-Emerson J, Kwan E, Besser TE, Cookson A, Carter PE, French NP. Whole-genome comparison of two Campylobacter jejuni isolates of the same sequence type reveals multiple loci of different ancestral lineage. PLoS One. 2011;6:e27121. doi: 10.1371/journal.pone.0027121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krauland MG, Dunning Hotopp JC, Riley DR, Daugherty SC, Marsh JW, Messonnier NE, Mayer LW, Tettelin H, Harrison LH. Whole genome sequencing to investigate the emergence of clonal complex 23 Neisseria meningitidis serogroup Y disease in the United States. PLoS One. 2012;7:e35699. doi: 10.1371/journal.pone.0035699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bidossi A, Mulas L, Decorosi F, Colomba L, Ricci S, Pozzi G, Deutscher J, Viti C, Oggioni MR. A functional genomics approach to establish the complement of carbohydrate transporters in Streptococcus pneumoniae. PLoS One. 2012;7:e33320. doi: 10.1371/journal.pone.0033320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haft DH, Selengut JD, White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31:371–373. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denielou YP, Sagot MF, Boyer F, Viari A. Bacterial syntenies: an exact approach with gene quorum. BMC Bioinformatics. 2011;12:193. doi: 10.1186/1471-2105-12-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novichkov PS, Ratnere I, Wolf YI, Koonin EV, Dubchak I. ATGC: a database of orthologous genes from closely related prokaryotic genomes and a research platform for microevolution of prokaryotes. Nucleic Acids Res. 2009;37:D448–D454. doi: 10.1093/nar/gkn684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Byrne KP, Wolfe KH. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seret ML, Baret PV. IONS: Identification of orthologs by neighborhood and similarity-an automated method to identify orthologs in chromosomal regions of common evolutionary ancestry and its application to hemiascomycetous yeasts. Evol. Bioinform. Online. 2011;7:123–133. doi: 10.4137/EBO.S7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davidsen T, Beck E, Ganapathy A, Montgomery R, Zafar N, Yang Q, Madupu R, Goetz P, Galinsky K, White O, et al. The comprehensive microbial resource. Nucleic Acids Res. 2010;38:D340–D345. doi: 10.1093/nar/gkp912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madupu R, Brinkac LM, Harrow J, Wilming LG, Bohme U, Lamesch P, Hannick LI. Meeting report: a workshop on Best Practices in Genome Annotation. Database. 2010;2010:baq001. doi: 10.1093/database/baq001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purushe J, Fouts DE, Morrison M, White BA, Mackie RI, Coutinho PM, Henrissat B, Nelson KE. Comparative genome analysis of Prevotella ruminicola and Prevotella bryantii: insights into their environmental niche. Microb. Ecol. 2010;60:721–729. doi: 10.1007/s00248-010-9692-8. [DOI] [PubMed] [Google Scholar]