Abstract
Ammonia-oxidizing archaea (AOA) are thought to be among the most abundant microorganisms on Earth and may significantly impact the global nitrogen and carbon cycles. We sequenced the genome of AOA in an enrichment culture from low-salinity sediments in San Francisco Bay using single-cell and metagenomic genome sequence data. Five single cells were isolated inside an integrated microfluidic device using laser tweezers, the cells' genomic DNA was amplified by multiple displacement amplification (MDA) in 50 nL volumes and then sequenced by high-throughput DNA pyrosequencing. This microscopy-based approach to single-cell genomics minimizes contamination and allows correlation of high-resolution cell images with genomic sequences. Statistical properties of coverage across the five single cells, in combination with the contrasting properties of the metagenomic dataset allowed the assembly of a high-quality draft genome. The genome of this AOA, which we designate Candidatus Nitrosoarchaeum limnia SFB1, is ∼1.77 Mb with >2100 genes and a G+C content of 32%. Across the entire genome, the average nucleotide identity to Nitrosopumilus maritimus, the only AOA in pure culture, is ∼70%, suggesting this AOA represents a new genus of Crenarchaeota. Phylogenetically, the 16S rRNA and ammonia monooxygenase subunit A (amoA) genes of this AOA are most closely related to sequences reported from a wide variety of freshwater ecosystems. Like N. maritimus, the low-salinity AOA genome appears to have an ammonia oxidation pathway distinct from ammonia oxidizing bacteria (AOB). In contrast to other described AOA, these low-salinity AOA appear to be motile, based on the presence of numerous motility- and chemotaxis-associated genes in the genome. This genome data will be used to inform targeted physiological and metabolic studies of this novel group of AOA, which may ultimately advance our understanding of AOA metabolism and their impacts on the global carbon and nitrogen cycles.
Introduction
Mesophilic Crenarchaea (first discovered by Fuhrman et al. [1] and DeLong [2]) are among the most abundant microorganisms on the planet [1]–[2]. Crenarchaea constitute ∼10–40% of the total microbial community in the ocean below the euphotic zone [3]–[6]. In fact, it is estimated that there are 1.0×1028 crenarchaeal cells in the world's oceans compared to 3.1×1028 bacterial cells [3]. In soil systems, Crenarchaea generally comprise 1–5% of the total prokaryotic community [7]–[9], but can reach upwards to 38% in some systems (e.g., temperate acidic forest soils) [10]. It appears that the vast majority of Crenarchaea in soils and the open ocean possess ammonia monooxygenase (amoA) genes and thus may be capable of oxidizing ammonia to nitrite, based on quantitative PCR estimates of 16S rRNA and amoA genes, fluorescent in situ hybridization, and metagenomic analyses [11]–[18]. Recent phylogenetic and genomic analyses suggest that these mesophilic Crenarchaea should be recognized as a third archaeal phylum, the Thaumarchaeota [19]–[20].
AOA have proven difficult to isolate in culture, as evidenced by the fact that only one pure culture [21] has been documented despite reports of several enrichment cultures [12], [22]–[25]. Many questions remain about the physiology, metabolic and biogeochemical functions, evolutionary history, and ecological niche partitioning of AOA. Genome sequencing is a powerful tool that can be used to address these outstanding questions, yet traditional sequencing approaches require pure cultures or enrichment to near homogeneity. Genome sequencing of individual cells presents a new approach for exploring the genetic makeup of microorganisms present in heterogeneous mixtures (such as the enrichment culture that was the basis for this study) without the need for cell viability or growth. Single-cell genomics can be used independently on uncultured microbes [26]–[27] or as a complement to metagenomic studies on enrichment cultures and environmental samples by aiding in sequence binning and genome reconstruction [28].
Here, we used a combination of single-cell genomic and metagenomic approaches to sequence the genome of a novel, low salinity-type AOA. The single-cell approach is useful for obtaining organism-specific sequence datasets from heterogeneous samples, and furthermore provides an insight into micro-heterogeneity at the population level. The genomic data reveal sequence features not observed in other AOA, as well as similarities to published AOA genomes, which speak to the metabolic potential and environmental relevance of these organisms.
Results and Discussion
Enrichment of ammonia-oxidizing archaea from low-salinity sediments
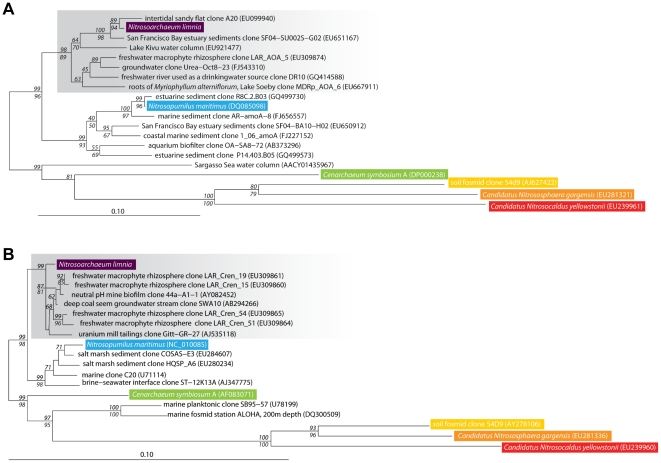
An ammonia-oxidizing enrichment culture initiated from sediments in the low-salinity region of San Francisco Bay was dominated by AOA; CARD-FISH showed that Archaea (Arch915 probe) accounted for approximately 84% of cells in the enrichment and the remaining 16% were Bacteria (Eub338 I–III probes). The AOA grew chemoautotrophically by aerobic ammonia oxidation to nitrite. Archaeal amoA and 16S rRNA gene sequences from the AOA enrichment culture were phylogenetically distinct (bootstrap supported) from N. martimus [Figure 1], and were instead most closely related to environmental clones derived from freshwater ecosystems. In total, over 500 GenBank amoA sequences fell within this phylogenetic cluster, 79% of which were from low salinity habitats. Another 7% of the sequences were from habitats that experience low salinity at some point in the year (e.g., San Pablo Bay sites in San Francisco Bay), and 4% of the sequences were from soil. Most of the remaining sequences were from salt marsh sediments [29], suggesting that this AOA phylotype may tolerate elevated salinity. Nevertheless, the vast majority of amoA sequences within this clade were from freshwater habitats.
Figure 1. Phylogeny of Ammonia Oxidizing Archaea gene sequences.
Neighbor-joining phylogenetic tree of (A) archaeal amoA gene sequences and (B) archaeal 16S rRNA gene sequences. Grey boxes highlight the putative low-salinity group. Significant bootstrap values (≥50) from 1000 replicates are shown in italics at branch nodes. Neighbor-joining bootstrap values are above the line and parsimony values are below the line.
On the basis of these results and the genome analysis described herein, we propose the provisional classification of this novel archaeon as Candidatus Nitrosoarchaeum limnia SFB1. The name refers to low-salinity habitat of this archaeon and to its ability to oxidize ammonia to nitrite. Nitrosoarchaeum comes from the Latin masculine adjective ‘nitrosus’ for nitrous and limnia is derived from the Greek word ‘limne’ for freshwater (as in limnology) and the strain name SFB1 refers to its isolation from San Francisco Bay. The short description of Candidatus Nitrosoarchaeum limnia is a mesophilic, chemolithoautotrophic ammonia-oxidizing archaeon belonging to the Thaumarchaeota phylum.
On-chip cell sorting
Based on this information suggesting that N. limnia is novel and distinct from N. maritimus, we employed a combined single cell sorting and metagenomic approach to obtain the genome sequence of these AOA. A variety of single-cell approaches to microbial genomics are currently being applied [26]–[27], [30]–[31]. While all rely on the multiple displacement amplification (MDA) reaction [32] for the amplification of unknown genomic DNA sequences, the means by which individual cells are isolated distinguish the various approaches. Microfluidic flow [30], flow cytometry [31], [33], and micromanipulation [27] have all been successfully applied to isolate single cells for genomic amplification, although each approach has strengths and weaknesses. In particular, microfluidics and micromanipulation allow microscopic examination of the sorted cells, while flow cytometry provides higher raw throughput. A challenge in this case was the tiny, submicron size of the AOA in the enrichment culture.
We implemented laser tweezer [34]–[35] based cell sorting inside a microfluidic device to combine the benefits of a sealed, integrated system for sorting and amplification, small reaction volumes, and minimum sorting volume. The imaging and trapping system was adapted to enable visualization and trapping of tiny cells. This required the application of a high-numerical aperture phase-contrast objective with high infrared transmission. All reagents were carefully filtered to remove extraneous particles that could be confused with cells. These features of the optical sorting method specifically and effectively address the three classes of contamination with the potential to compromise single cell amplification reactions, namely: the laboratory environment, amplification reagents, and cells/DNA from the sample itself.
Using this microfludic and laser trapping device, single cells smaller than one micron in length were sorted from the AOA enrichment culture [Figure S1]. The cells were lysed ‘on-chip’ using proteinase K and alkaline lysis, and the genomes of each individual cell were amplified by MDA. Amplified cells were screened for archaeal amoA and 16S rRNA genes to confirm affiliation with AOA. Based on PCR screening, we selected the MDA products of five individual AOA cells for genomic sequencing. In addition, due to the highly enriched nature of the ammonia-oxidizing culture (with respect to the AOA), we also carried out MDA on a benchtop scale using cells from the enrichment culture as the template. We directly sequenced the enrichment MDA products to obtain sequences from the ‘bulk’ enrichment culture metagenome. This combined single cell/metagenomic approach enabled us to test the single cell assemblies for completeness and the metagenome for contamination.
Sequencing and assembly
MDA-amplified genomic DNA from the five individual AOA cells was sequenced using a combination of shotgun (SG) and paired-end (PE) DNA pyrosequencing approaches on the Roche/454 FLX Titanium platform. MDA products from both individual cells and the bulk enrichment were used to create sequencing libraries, which were quantitated using digital PCR [36] and sequenced. We analyzed 483,350 SG and 56,122 PE reads (119 Mb) obtained from five individual cells and 160,632 SG and 18,118 PE reads (37 Mb) from the enrichment sample [Table 1]. The G+C contents of the single-cell reads formed a major peak centered at 32.4%, an even lower G+C content than N. maritimus (34.2%) [Figure S2]. To compare the dispersion of the G+C content, we simulated shotgun reads from N. maritimus with the same average read length as the experimentally derived datasets. The dispersion of the read G+C content from the single-cell reads was essentially identical to that found of N. maritimus, confirming that the individual AOA cells were amplified without contamination from bacterial cells or DNA. The G+C content of the enrichment reads demonstrated a principal mode similar to the single cell datasets and N. maritimus (suggesting a large proportion of AOA in the enrichment), as well as a higher G+C component attributed to sequences from the bacteria present in the culture.
Table 1. N. limnia assembly statistics for the consensus genome (metagenome and five single cells), single cell coassembly (five single cells), and three individual cell assemblies.
| assembly statistic | Consensus | Single cells only | Cell 90507.23 | Cell 90507.21 | Cell 90507.3 |
| raw reads | 693,589 | 530,748 | 70,156 | 200,938 | 138,886 |
| paired-end reads | 33,725 | 26,953 | 3,018 | 517 | 2,388 |
| raw bases | 150,994,537 | 118,796,782 | 17,107,411 | 52,341,561 | 29,999,202 |
| assembly bases | 1,769,573 | 1,690,404 | 1,094,113 | 1,039,820 | 1,041,604 |
| number of elements | 31 | 136 | 355 | 253 | 348 |
| number of scaffolds | 2 | 26 | 68 | 76 | 83 |
| scaffold bases | 1,757,035 | 1,366,809 | 852,837 | 892,230 | 834,370 |
| scaffold N50 | 1,363,261 | 155,965 | 20,543 | 16,443 | 14,147 |
| largest scaffold | 1,363,261 | 263,850 | 70,423 | 49,773 | 41,297 |
| contig bases | 1,740,160 | 1,664,128 | 1,090,610 | 1,039,764 | 1,031,668 |
| contig N50 | 94,192 | 28,182 | 14,827 | 14,834 | 9,613 |
| unscaffolded contigs | 29 | 110 | 287 | 177 | 265 |
| largest contig | 230,803 | 135,296 | 70,423 | 49,773 | 27,947 |
Individually, three of the single-cell datasets gave assemblies in excess of one megabase from sequencing depths of 10× to 30×, based on an expected genome size of about 1.7 Mb. The implied 60% genomic coverage is typical in our experience with single-cell genomic data at such depths of sequencing. The limited genomic coverage of the single-cell assemblies owes to the phenomenon of MDA amplification bias, wherein certain regions of the genome are amplified to a greater extent than other regions. The less-amplified regions are not efficiently sampled by the shotgun sequencing procedure, and extremely high sequence coverage is required to recover the under-represented sequences.
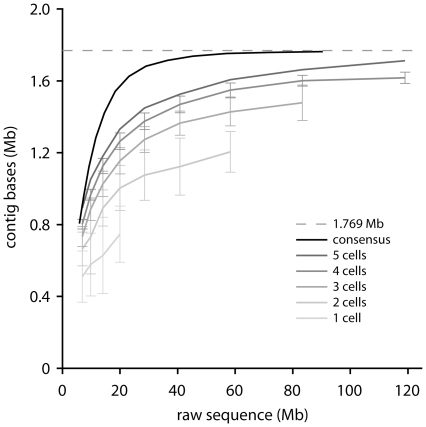
Interestingly, each of these single-cell assemblies represents a different 60% of the target genome, indicating that the genomic coverage and/or amplification bias profile differs from cell to cell. This phenomenon is consequential for single-cell genomic analyses, and can be utilized to improve study outcomes when considered vis a vis project goals at the stages of cell screening, DNA sequencing, assembly and analysis. Specifically, the lack of correlation among the amplification profiles of independently amplified cells allows the improvement of coverage profiles by combining datasets from different individual cells. Thus, while further sequencing and efforts to improve the assembly suffer diminishing returns with respect to an individual cell, great strides in assembly quality are quickly won by combining the reads obtained from several individual cells. By pooling the data from the five individual cells sequenced, we obtain an assembly of 136 elements (26 scaffolds and 110 contigs) for a total of 1.69 Mb of sequence, 1.37 Mb of which was captured in the scaffolds (scaffold N50 = 156 kb) [Table 1]. A combination of rarefaction analysis [Figure 1] and comparison to the metagenomic data indicates (Table 1) that this single-cell assembly represents >95% of the N. limnia genome.
The metagenome from the bulk enrichment was used as an independent validation of the single cell data and to supplement the single cell analysis. To combine the strengths of each dataset—the pure single cell AOA reads with no bacterial sequences and the bulk enrichment reads with less amplification bias—we generated a high-quality consensus assembly. First, we used five single-cell datasets to establish a stringent G+C content filter and eliminate high G+C content (>46%) reads from the enrichment dataset, purging much of the bacterial sequence contribution prior to assembly of the data. Next, we mapped the single-cell reads to a de novo assembly of the filtered enrichment reads to identify and reject chimeric sequences, the occurrence of which is expected to be independent in different datasets. The small pool of high-G+C reads was mapped against N. maritimus and C. symbiosum A to recover high G+C content conserved sequences. The consensus assembly contained 2 scaffolds and 29 contigs for a total of 1.77 Mb (contig N50 = 94 kb) [Table 1]. More than 99% of the assembled bases were represented by the two scaffolds (1.36 Mb and 0.39 Mb in length). The consensus assembly represents greater than 97% of the genome, based on rarefaction analysis. The elements from both assemblies were oriented and ordered by homology to N. maritimus. Orientation was determined by maximizing the sum of the alignment score to this reference, and the elements ordered according to the hit length-weighted average subject position of the homologous regions that were co-orientated with the reference. The genome sequence is reported in the supplementary information file Data S1.
Microheterogeneity and sequencing (MDA) error
Comparing homologous regions of the single cell assemblies with each other and with the enrichment dataset, we consistently find 99.9% average nucleotide identity. The identity result is a lower limit because of the possibility of errors introduced in the amplification and sequencing procedures. Newbler reports 5904 bases in the assembly as having quality scores lower than 40. If we conservatively estimate the average quality score of these lower-quality aligned bases at 10 and the average quality score of the high-quality aligned bases at 40, 234 errors arising from ambiguous alignments and low-quality raw bases are predicted. Based on independent measurements (data not shown), we expect a minor contribution of additional errors based on the MDA procedure, on the order of 1/100,000 bp, or fewer than 20 bases across the N. limnia genome. Thus, it seems that microheterogeneity among the enrichment AOA must account for a substantial portion of the SNPs predicted among the individual cells.
Genome size estimate
Subsets of the enrichment dataset were assembled in a rarefaction analysis to determine the approximate genome size of N. limnia [figure 2]. The total assembly size smoothly asymptotes below 1.80 Mb, indicating that the N. limnia assembly represents an essentially complete genome at 1.77 Mb. We also rarefied the single-cell data, which we expected to assemble less efficiently than the enrichment dataset, due to the effect of greater amplification bias in the single cell MDA reactions compared with the bulk MDA of the enrichment sample. This expectation was borne out, but the single-cell data (in aggregate) do asymptote at approximately the same level. This suggests that the single-cell dataset contains all the genomic sequences, albeit with less genomic information per raw Mb as a result of representation bias. Assembly using only the single cell data yields a genome that is estimated to be 95% complete [Table 1]. It is interesting to imagine the impact of sequencing additional cells.
Figure 2. Assembly of the N. limnia datasets.
Rarefaction analysis of the N. limnia sequence data showing assembly of the consensus dataset and as a function of the number of included single-cells.
Gene prediction and annotation
Gene prediction was carried out on the N. limnia assembly using the FgenesB platform (Softberry, Inc.), which called 2171 genes, including 2047 protein-coding ORFs, 74 pseudogenes, and 50 RNA genes [Table 2]. In total, 83.4% of the assembled bases were predicted to code for proteins. The N. limnia genome encodes 45 transfer RNA (tRNA) genes (42 identified by FgenesB and 3 additional tRNA sequences identified by comparison with N. maritimus). tRNAs for 18 amino acids were predicted, which further included two tRNAs with unidentified codon/amino acid specificity. Single copies of the full-length 16S, 23S, 5S, and 5.8S ribosomal RNA genes exist in the assembly. The average nucleotide identities of the 16S and 23S genes to N. maritimus are of 97% and 94%, respectively. In addition, a conserved noncoding RNA with 93% identity to N. maritimus was identified. The annotation data is reported in the supplementary information file Data S2.
Table 2. Genome features for N. limnia, N. maritimus, and C. symbiosum A.
| Specifications | N. limnia | N. maritimus | C. symbiosum A |
| Size, bp | 1,769,573 | 1,645,259 | 2,045,086 |
| Average G+C content, % | 32 | 34 | 58 |
| Predicted genes | 2,171 | 1,847 | 2,066 |
| Average gene length (bp) | 696 | 825 | 924 |
| Coding percentage, % | 83 | 90 | 91 |
| Gene content | |||
| Pseudogenes | 74 | 6 | 0 |
| Total number of ORFs | 2,047 | 1,797 | 2,017 |
| Predicted functional | 1,136 | 1,082 | 994 |
| Hypotheticals | 911 | 714 | 1,023 |
| ORF density (ORF/kb) | 1.16 | 1.09 | 1.01 |
| RNA genes | |||
| 16S-23S rRNA operon | 1 | 1 | 1 |
| 5S rRNA | 1 | 1 | 1 |
| tRNAs | 45 | 44 | 45 |
| other RNA | 2 | 2 | 1 |
Codon usage and nucleotide skew
The N. limnia codon usage was compared with that of N. maritimus and C. symbiosum A, with a nucleotide-matched scrambled control as a reference (data not shown). The codon usage of N. limnia is very similar to that of N. maritimus, which has a similar G+C content, and distinct from both the high G+C content C. symbiosum A and the randomized control. This is also the case for N. limnia CDS lacking homology to database sequences, supporting the identification of these regions as protein-coding genes.
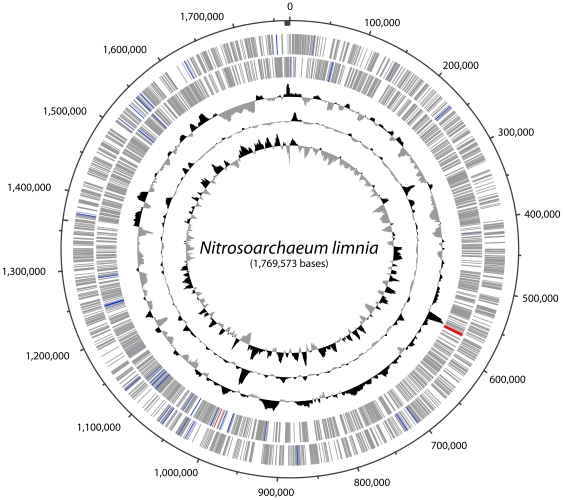
The G+C skew in the N. limnia genome exhibits an irregular pattern comparable to that observed in other archaeal genomic sequences [figure 3] [37]. Analysis of cumulative G+C and GGG+T skew failed to reveal an obvious origin of replication, although several strong transition points exist in the genome.
Figure 3. Circular representation of the N. limnia genome.
From the outer ring to the inner ring: Scaffold breakpoints are indicated by the inside tick marks, predicted genes on the forward strand, predicted genes the reverse strand, G+C content, GC skew, and GGGT skew. On the outer two rings, protein coding sequences are colored gray, tRNA genes in blue, rRNA genes in red, and noncoding RNA genes in green.
Gene prediction and percent identity comparisons
The N. limnia genome has a coding density of 1.16 protein coding genes per kb of sequence data, which is slightly higher than the coding density for N. maritimus (1.09) or C. symbiosum A (0.99) [Table 2]. The N. limnia genome shares 1423 genes with N. maritimus and 1156 genes with C. symbiosum A (based on Reciprocal Best Hit [RBH] analysis). The 1423 genes that were RBHs with N. maritimus genes represent 66% of the genes predicted in the N. limnia assembly. Conversely, 75% of the N. maritimus genes were identified as RBHs in the N. limnia assembly. Only 53% of genes in the N. limnia assembly were RBHs with C. symbiosum A, despite the larger genome size of C. symbiosum A, indicating the greater evolutionary distance that exists between N. limnia and C. symbiosum A than exists between N. limnia and N. maritimus.
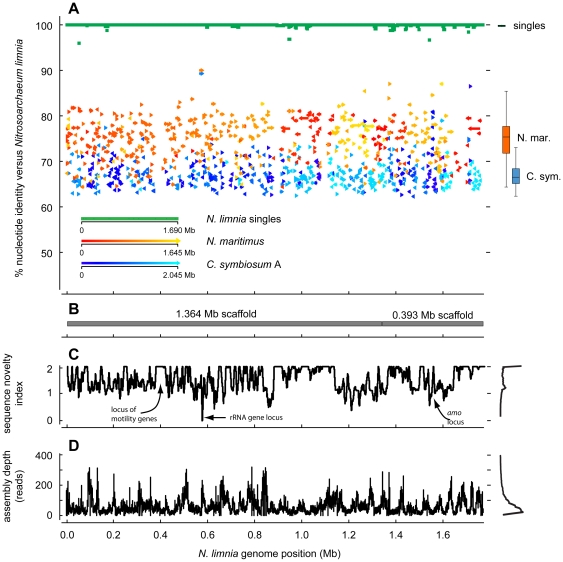
When compared across the entire length of the genome, N. limnia was distinct from both N. maritimus and C. symbiosum A [Table 3 and Figure 4]. The average nucleotide identity across overlapping regions of the N. limnia and N. maritimus genomes was 76.3% [Table 3]. When scoring non-overlapping genomic regions at an arbitrary 25% nucleotide identity, the overall nucleotide identity declines to 68.1%. The percent identity between the N. limnia and C. symbiosum A genomes was even lower, as expected due to the divergent total nucleotide content: 67.4% identity across overlapping regions and 33.3% when including non-overlapping regions [Table 3]. The cutoff for defining a new species was recently proposed to be 95–96% average nucleotide identity between two genomes [38]—an updated method of species identification compared to DNA-DNA hybridization. Thus, the much lower percent identity between N. limnia, N. maritimus and C. symbiosum A across the entire genome supports the classification of N. limnia as a new genus of AOA. The N. limnia and N. maritimus genomes shared extensive regions of synteny (gene order), whereas N. limnia and C. symbiosum A showed very little synteny across the genomes [Figure 4 and Figure S3].
Table 3. Whole genome nucleotide identity comparisons between N. limnia, N. maritimus, and C. symbiosum A.
| Whole genome comparison | N. maritimus | C. symbiosum A |
| number of bases in corresponding regions | 1,180,862 | 398,085 |
| % identity of corresponding regions | 76.3% | 67.4% |
| whole genome average % identity* | 68.1% | 33.3% |
*given 25% identity in non-corresponding regions
Figure 4. Comparison of three AOA genomes.
(A) The top panel shows nucleotide identity of blast hits between the reference genomes and the N. limnia genome. Hits for N. maritimus and C. symbiosum A are colored according to the position in the reference (query) genome. The arrow direction indicates the hit direction. On the right, box plots summarize the distribution of hits from each reference genome to the N. limnia genome. (B) The positions of the two largest consensus scaffolds are shown. (C) The sequence novelty index, defined as 2 minus the sum of nucleotide identity of blast hits to N. maritimus and C. symbiosum A at each position is plotted. A histogram of the values is present at right. (D) The alignment depth in the final assembly is shown. A histogram of the values is present at right.
Motility
One of the most striking features of the N. limnia genome, relative to N. maritimus and C. symbiosum, is the presence of numerous motility-associated genes, including those required for Flp pilus assembly, flagellar assembly, and chemotaxis [Table 4]. In total, N. limnia has 38 motility-associated genes, many of which are clustered in a locus on the largest scaffold [Figure 4]. This is a larger number than observed in many other archaea including Sulfolobus solfataricus, Aeropyrum pernix, and Archaeoglobus fulgidus, which all have fewer predicted motility genes (15–20 each). The N. limnia flagellar genes share homology with thermophilic Crenarchaeota. In addition, the majority of the cells in the AOA enrichment culture were observed to be motile by optical microscopy. This appears to be a distinctive trait from N. maritimus and C. symbiosum A, both of which lack gene suites for assembly of flagella. N. limnia was isolated from estuarine sediments, raising the possibility that this AOA requires motility to position itself in response to varying substrate and/or oxygen concentrations in the surface sediments. In contrast, N. maritimus was isolated from gravel in a tropical fish tank and C. symbiosum A is a symbiont of a marine sponge, where perhaps motility is less essential. Determining how widespread motility and chemotaxis are within phylogenetically diverse AOA genera (from water columns, soils, sediments, etc.) represents an exciting new avenue of research.
Table 4. N. limnia genes putatively associated with motility.
| Motility Associated Genes | Number of Genes |
| Flagella: | |
| Archaeal flagella assembly protein J | 1 |
| Archaeal flagellins | 5 |
| Predicted ATPases involved in biogenesis of archaeal flagella | 1 |
| Type IV secretory pathway, VirB11 components, and related ATPases involved in archaeal flagella biosynthesis | 1 |
| Chemotaxis: | |
| Chemotaxis protein histidine kinase and related kinases | 2 |
| Chemotaxis protein; stimulates methylation of MCP proteins | 1 |
| Chemotaxis response regulator containing a CheY-like receiver domain and a methylesterase domain | 2 |
| Chemotaxis signal transduction protein | 1 |
| CheY-like receiver | 19 |
| Methyl-accepting chemotaxis protein | 2 |
| Methylase of chemotaxis methyl-accepting proteins | 1 |
| Pilus: | |
| Flp pilus assembly protein TadC | 2 |
Protection from osmotic shock
The N. limnia genome contains one large-conductance mechanosensitive channel protein and two small-conductance mechanosensitive channel proteins. Mechanosensitive (MS) channels are found in bacteria, archaea, and eukaryotes. MS channel proteins protect cells from hypoosmotic shock when a cell transitions from a high osmolarity environment to a low osmolarity environment [39]. This transition causes a rapid influx of water into the cells, which increases the internal turgor pressure and places considerable strain on the cytoplasmic membrane and cell wall. To release this pressure, the MS channels open, allowing the release of low molecular mass solutes (K+, glutamate and compatible solutes) out of the cell. This rapidly decreases the concentration of osmotically-active solutes in the cytoplasm, minimizing the influx of water and thereby preventing cell lysis. The C. symbiosum A and N. maritimus genomes appear to lack large-conductance MS channels, although they each encode at least one small-conductance MS channel protein. N. limnia was isolated from sediments in the San Francisco Bay estuary that likely experience regular fluctuations in salinity and may depend on these MS channel proteins for protection from osmotic shock.
The N. maritimus genome contains genes coding for ectoine biosynthesis, representing the first description of this process in Archaea [40]. Bacteria and Archaea accumulate many different organic osmolytes, including ectoine, in response to high osmotic pressure [41]. Notably, the N. limnia genome is missing all four genes for ectoine biosynthesis (ectA, ectB, ectC, and ectD). Although the N. limnia genome is not closed, the fact that the genome is missing all four genes suggests that N. limnia does not use ectoine biosynthesis as a mechanism for dealing with osmotic stress. These genomic differences may in turn reflect niche differentiation between N. maritimus (marine) and N. limnia (low-salinity).
Genes conferring resistance
Several phage integrase proteins were identified in the N. limnia genome. However, no obvious CRISPRs (clustered regularly interspaced short palindromic repeats) were detected in the genome, which confer resistance to phage infection in many archaeal species, despite the fact that CRISPRs are found within ∼90% of archaeal genomes [42] and references within]. Thus, N. limnia may martial other defenses—the genome codes for at least one restriction-modification systems—or may be particularly susceptible to phage infection. Three transposase genes were also present in the genome and may be an additional source of genetic exchange and rearrangement. Several genes putatively conferring antibiotic resistance were found in the N. limnia genome (e.g. beta-lactamase domain-containing proteins and an antibiotic biosynthesis monooxygenase). The presence of several metal transporters and proteins implicated in metal resistance may reflect metal exposure in the San Francisco Bay sediments; indeed, there are many potentially toxic metals in the sediments of North San Francisco Bay [23].
Novel cell division system
In Euryarchaeota and Korarchaeota (as well as most bacteria), cell division is mediated by FtsZ proteins, which are homologous to tubulin, the building block of the microtubule cytoskeleton in eukaryotes [43]–[44]. The sequenced genomes of hyperthermophilic Crenarchaeota lack FtsZ genes. Instead, it appears that all Crenarchaeota (with the exception of the Thermoproteales, where the cell division machinery remains uncharacterized) use a novel Cdv cell division system that is related to the related to the eukaryotic ESCRT-III sorting complex [44]–[45]. However, both the FtsZ and Cdv systems of cell division are evident in the AOA genomes, including N. limnia, N. maritimus, and C. symbiosum A, as well as the draft genome of Nitrososphaera gargensis [20]. The striking difference in cell division between these AOA and all other Crenarchaea supports the proposal that they belong to a third archaeal phylum, the Thaumarchaeota [19]–[20].
Chromosome structure and organization
Prokaryotic chromosomal DNA is highly compacted to form a nucleus-like structure—the nucleoid. Bacteria and Archaea have to compact their genomic DNA by over 1000-fold [46] by utilizing a wide variety of DNA binding chromatin proteins. These proteins not only serve to compact the genomic DNA, but also in maintaining the genome in a state that allows access for gene expression, DNA replication and repair. The two major archaeal chromatin proteins are the archaeal histone and Alba proteins. Archaeal histones act as spools that DNA wraps around (similar to their eukaryotic homologs), whereas Alba proteins form extended filamentous fibers containing DNA [47]. All archaea have either one of these proteins, and many encode both proteins [48]. The N. limnia genome codes for one histone protein (Nlim1809) and two Alba proteins (Nlim0610 and Nlim1213).
Carbon metabolism
The N. limnia genome sequence suggests that this organism uses a modified version of the 3-hydroxypropionate/4-hydroxybutyrate pathway for carbon fixation, as in N. maritimus and C. symbiosum A [37], [52]. In addition to pathways for autotrophic carbon fixation, genes putatively inferring the function of organic carbon consumption have been identified in the genomes of N. limnia, N. maritimus and C. symbiosum A. Several isotopic studies have suggested that natural populations of AOA may be capable of mixotrophic growth [4]–[5], [49]–[51]. It remains to be determined how important heterotrophy and/or mixotrophy is to AOA in natural environments.
Ammonia oxidation and respiratory chain
In ammonia-oxidizing bacteria (AOB), ammonia is oxidized to hydroxylamine by ammonia monooxygenase (AMO) composed of α, β, and γ subunits encoded by the amoA, amoB, and amoC genes, respectively. Hydroxylamine oxidoreductase (HAO) then oxidizes hydroxylamine to nitrite, generating four electrons that are transferred to the terminal oxidase and AMO via cytochromes c554 and c M552 and the ubiquinone pool ([52], [53] and references therein).
The N. limnia genome contains genes putatively encoding the α, β, and γ subunits of the archaeal AMO protein. The gene order in the AMO operon is conserved amongst N. limnia, N. maritimus [40], and the Sargasso Sea metagenomic fragments [54]: amoA, hypothetical protein, amoC, followed by amoB. The N. limnia AMO gene sequences have relatively low identities to N. maritimus, ranging from 80–91% nucleotide identity and 77–96% amino acid identity [Table 5].
Table 5. Nucleotide (NT) and amino acid (AA) identities for key individual genes in N. limnia, N. maritimus, and C. symbiosum A.
| N. limnia vs | N. limnia vs | |||
| N. maritimus | C. symbiosum A | |||
| Individual gene comparison | AA Identity | NT Identity | AA Identity | NT Identity |
| 16 s rRNA | n/a | 97% | n/a | 94% |
| 23 s rRNA | n/a | 94% | n/a | 89% |
| ammonia monooxygenase subunit A | 95% | 89% | 95% | 78% |
| ammonia monooxygenase subunit B | 92% | 88% | 76% | 75% |
| ammonia monooxygenase subunit C | 96% | 91% | 93% | 81% |
| AMO-associated hypothetical protein ** | 77% | 80% | 73% | 72%*** |
**ammonia monooxygenase operon-associated hypothetical protein
***<90% sequence coverage
While the details of the ammonia oxidation and electron transfer pathways remain to be elucidated, AOA, including N. maritimus, C. symbiosum A and N. limnia, appear to have an ammonia oxidation pathway distinct from AOB [40],[55]–[56]. The N. maritimus, C. symbiosum A and N. limnia genomes lack homologues for HAO or cytochromes c554 and c M552, and it remains to be demonstrated whether hydroxylamine is an intermediate in the pathway. AOA in pure culture produce nitrite from the oxidation of ammonia [21], [57] but the genome sequence annotations do not include the key HAO enzyme necessary to produce nitrite. The archaeal AMO protein may not produce hydroxylamine (explaining the missing HAO protein) or the AOA may contain novel enzymes responsible for the oxidation of hydroxylamine. In either case, redox reactions in AOA are likely more reliant on copper than iron as seen in the AOB with the iron-rich HAO enzyme and cytochromes [40], [55]. The three AOA genomes all contain numerous copper-containing proteins, including multicopper oxidases (MCOs) and small blue copper-containing proteins. These predicted copper-containing proteins might form the basis of electron transfer in AOA with functionality similar to cytochromes in AOB [40], [55]. The N. limnia genome contains 11 blue (type 1) copper domain-containing proteins, 1 cupredoxin blue copper protein, and 4 multicopper oxidases [Table S1]. Alternative pathways for ammonia oxidation have been proposed by Walker et al. [40]. The N. limnia genome does not further illuminate the exact mechanism for ammonia oxidation.
Two ammonium transporter genes were identified in the N. limnia genome (Nlim1421 and Nlim1564). If the AMO protein accesses ammonia outside of the cytoplasmic membrane (as in AOB), the AOA would not necessarily need ammonium transporters. However, the RBHs exist between the N. limnia ammonium permeases and both N. maritimus and C. symbiosum A. The existence of conserved ammonium transporters in all three genomes suggests a necessary function for this activity. Arp et al. (2007) [52] proposed that ammonium transporters in AOB might facilitate the regulation of AMO and HAO that is induced by ammonium or to assist in the uptake of ammonia for nitrogen assimilation.
Many AOB are able to use urea as an alternative source of ammonia and carbon dioxide for growth [58]. Ureolysis may therefore be an ecological advantage in environments with low pH and/or fluctuating ammonia and carbon dioxide availability. Like N. maritimus [40], there was no evidence that N. limnia has the ability to use urea as an alternative source of ammonia, because no genes encoding urea transporters or urease enzymes were identified in the genome. Interestingly, however, both urea transporter and urease genes were present in the C. symbiosum A genome [55]. The symbiotic C. symbiosum A may use ureolysis as a mechanism to remove nitrogenous waste from its host, the marine sponge Axinella mexicana [56]. Konstantinidis et al. [16] also reported urease genes in a crenarchaeal genomic scaffold from 4,000-m-depth at station ALOHA in the Pacific Ocean (along with amoA, amoB, amoC, and ammonia permease genes), suggesting that urease activity is a feature of at least some marine water column AOA.
Conclusion
We used an approach combining metagenomic and single cell data to produce a high quality and complete (but not fully closed) genome of a mesophilic AOA from a low-salinity estuarine environment – Nitrosoarchaeum limnia. Single-cell genome sequencing enabled discovery and assembly of 95% of the complete N. limnia genome; when this data was combined with metagenomic reads derived from the bulk enrichment culture, greater than 97% of the genome was determined. The genomic sequence reflects several systems present in the two other AOA genome sequences available, including the machinery for cell division and ammonia oxidation, establishing common elements that define this functionally related group of archaea belonging to the proposed archaeal phylum Thaumarchaeota. The N. limnia genome also reveals features unique to this environmental AOA, including a substantial suite of genes for flagellar biosynthesis and chemotaxis, and the presence of a large mechanosensitive channel that may be crucial for fitness in the estuarine environment. Several large regions of the genome lacking homology to N. maritimus and C. symbiosum may encode other functional genes important to the survival of free-living, estuarine AOA. Cultivation and genomic sequencing of environmentally-relevant AOA portends further physiological and metabolic studies of this novel and pervasive group of organisms that will ultimately advance our understanding of the global carbon and nitrogen cycles.
Materials and Methods
Cultivation of archaeal ammonia-oxidizers
Ammonia-oxidizer enrichment cultures were initiated from sediments collected in northern part of the San Francisco Bay estuary in 2006 (site SU001S; latitude 38°5′55.75″N, longitude 122° 2′47.40″W). Surface sediments were inoculated into a modified low-salinity version of Synthetic Crenarchaeota Medium [21], as described by Mosier and Francis (2008) [23]. Cultures were grown in static culture flasks or serum vials in the dark at room temperature (approximately 22°C). Approximately 10% of the culture volume was routinely transferred into fresh media.
The presence of ammonia-oxidizing archaea was confirmed by gene sequencing and fluorescent in situ hybridization (FISH). Briefly, DNA was extracted from the enrichment culture and archaeal 16S rRNA (Arch21F and Arch958R primers; [1]) and archaeal amoA (Arch-amoAF and Arch-amoAR primers; [59]) genes were PCR amplified, cloned and sequenced. Catalyzed reporter deposition fluorescence in situ hybridization (CARD-FISH; e.g., [60]) was carried out with horseradish peroxidase (HRP)-labeled probes Arch915 [61] and EUB338 I–III [62]–[63]. Filters were treated with lysozyme or proteinase K for permeabilization of bacteria and archaea, respectively.
Phylogenetic trees
Archaeal amoA nucleotide sequences were aligned with GenBank sequences using MacClade version 4.08 [64]. Archaeal 16S rRNA gene sequences were aligned using the NAST alignment server [65]. Sequences were then manually adjusted and aligned to the Greengenes 16S rRNA gene database [66] in ARB [67]. For phylogenetic analyses, a 582-bp region of the sequence alignment was used for amoA and a 1272-bp region for 16S rRNA. Neighbor joining (based on Jukes-Cantor correction) and parsimony phylogenetic trees were constructed in ARB [67] with 1000 bootstrap replicates. The set root option in ARB was used to manually root the trees between the sediment and water column cluster sequences.
Microfluidic device and cell sorting
Microfludidic devices with 32 units for nanoliter MDA reactions were produced by the Stanford microfluidics foundry. These devices are similar to those described in Marcy et al. [26]. The chip was pre-treated for 10 minutes with pluronic F127 at 0.2% in 1× PBS before filling with 1x PBS containing 0.01% Tween-20 and 0.01% pluronic F127 to reduce cell adhesion. The sample was pre-treated with 0.5–1 µg/mL proteinase K at room temperature for 10 minutes and then washed in 1× PBS and resuspended in 50∶50 1× PBS∶Ethanol. BSA was added to the treated cells (0.1 mg/mL final concentration). Individual cells were sorted one at a time from the bulk sample using the laser trap, through a two-valve gate, opening one valve at a time to allow the trapped cell, but not fluid flow, to pass through (see Movie S1). Each trapped cell was moved about 1 mm from the bulk sample to the reaction chamber using the laser trap.
Single-cell amplification
The Repli-G midi MDA reagents (Qiagen) were used to amplify individual cells in 50–60 nL volumes on the device, yielding 107–108 genomic equivelants of dsDNA product. First, cells contained in 0.75 nL PBS with 0.02% Tween-20 were flushed into the first lysis chamber with 3 nL buffer DLB (supplemented with 0.1 M dithiothreitol) to complete lysis and denature the genomic DNA. Then, ∼50 nL of reaction mix (45 µL were prepared from 29 µL Repli-G reaction buffer, 10 µL 20 mM H2O with 0.6% tween, 2 µL Repli-G enzyme, and 2 µL Repli-G stop solution) was added to each of the 32 reactions. The chip was then transferred to a hot plate set to 32°C and incubated overnight. This was carried out by fitting the outboard product ports on the chip with plastic pipet tips (P10 size), and by flushing the products into the pipet tips with the TRIS solution plumbed into the reagent port at 8 psi.
Six µl of each first-round reaction product were re-amplified using the Repli-G midi kit (Qiagen). Additionally, 1 µl of the proteinase K-treated cell solution used for sorting was amplified. The template solution was denatured by the addition of 3.5 µl buffer DLB for 5 minutes at room temperature. The solution was neutralized by adddition of 3.5 µl stop solution. A reaction mix consisting of 29 µl reaction buffer, 10 µl water, and 1 µl enzyme was prepared on ice, and then added to the denatured template. Reactions were incubated at 30°C for 12 hours and then diluted 10-fold in 10 mM TRIS with 0.02% Tween-20 and stored at −60°C.
Shotgun and paired-end library prep
Shotgun libraries were prepared from approximately 5 µg of the second round MDA product according to the Roche/454 protocol for “Titanium” shotgun libraries with the following modifications. Custom barcoded adaptor oligos (IDT) were used to enable pooling multiple libaries in a single emulsion PCR reaction and picotiter plate region during sequencing. To obtain dsDNA sequencing libraries and shorten the library preparation process, the library immobilization, Fill-in, and single-stranded library isolation steps (steps 3.7–3.9) were omitted.
Paired end libraries were prepared from approximately 5 µg of the second round MDA product according to the Roche/454 protocol for “Titanium 3 kb span” libraries with the following modifications. Custom barcoded adaptor oligos (IDT) were used in step 3.9.2 to enable pooling multiple libaries in a single emulsion PCR reaction and picotiter plate region during sequencing. To obtain dsDNA sequencing libraries and shorten the library preparation process, the library immobilization and single-stranded library isolation steps (steps 3.12.1 and 3.12.2) were omitted.
Sequencing library quantification
Sequencing libraries were quantified using digital PCR as previously reported [36], with the exceptions that 48.770 digital arrays (Fluidigm) were used for the microfluidic dPCR step, and that amplification primers complimentary to the Titanium adaptor sequences were used. Briefly, serial dilutions of the sequencing libraries were made in 10 mM TRIS buffer with 0.02% Tween-20. 48 sample preparations were then combined per the Fluidigm dPCR protocol with a reaction buffer containing thermostable DNA polymerase, dNTPs, GE sample loading reagent (Fluidigm), and the primers and probe necessary to carry out the universal Taqman amplification/detection scheme. The samples were loaded in the chip and run on the Biomark thermocycler for 45 cycles. Sample analysis was carried out using the default parameters for dPCR analysis using the Fluidigm analysis software. The quantitated libaries were then diluted to 2×106 molecules per microliter in 10 mM TRIS with 0.02% Tween-20 and aliquotted for storage at −60°C.
Genome sequencing
DNA pyrosequencing of the shotgun and paired-end libraries was carried out on the Roche 454 Genome Sequencer FLX instrument using “Titanium” chemistry. Emulsion PCR was carried out with DNA∶Bead ratios of 0.3∶1 for the shotgun libraries and 2.5∶1 for the paired-end libraries.
Sequence assembly
Reads from the shotgun and paired end pyrosequencing runs were separated by sample and trimmed using the sfffile tool (Roche) permitting one error in each 10 bp barcode. The G+C content of the single-cell reads formed a major peak centered at 32.4%, about 2% lower than the average G+C content of N. maritimus (34.2%). The dispersion of the read G+C content was essentially identical to that of N. maritimus, with the latter sampled at the same average read length. The few reads with G+C content greater than 46% were mapped to the N. maritimus genome, and mapping reads (90% identity over 40 bases) were combined with the low G+C content reads for subsequent analyses. The reads from the enrichment sample were then assembled de novo using Newbler (Roche). All assembly operations were carried out with Newbler v2.3 using default parameters. Paired end reads from the enrichment sample tagged by the assembler as ‘chimeric’ were excluded from subsequent operations. Reads from the single cells were then mapped to the initial enrichment assembly, and ‘chimeric’ reads excluded. Contigs made up of single-cell reads were then sampled as 550-mers with 50 base overlaps and included as reads in the final de novo assembly of the single-cell data. A second, ‘consensus’, de novo assembly was also perfomed using the single cell pyrosequencing reads (∼120 Mb) and the reads from the enrichment (∼37 Mb). A small number of short outlier contigs were excluded from the final assembly due to the identification of sequences from known contaminants from the MDA kit, such as Delftia acidovorans.
Gene annotation
Genes were predicted on the assembled genome sequence using the FgenesB program (Softberry, Inc.). Automated FgenesB annotations were manually refined by reviewing Reciprocal Best Hits (RBH) to Nitrosopumilus maritimus and Cenarchaeum symbiosum A and BLAST searches against the GenBank database. Individual proteins of interested were evaluated against “InterPro: the integrative protein signature database” [68] using InterProScan. RBH were identified using the blastp algorithm of Blast+ in conjunction with custom Matlab code. RBH are classified as “high confidence” when the forward hit covered at least 60% of the query sequence for the gene pair. Pseudogenes lacking a start or stop codon (74 in total) were included in the gene-based analyses. In total, 1200 genes were functionally annotated and 607 have strong homology to another organism but no assigned function. 323 unannotated genes (156 of which are 30 aa in length or less) were labeled as “hypothetical proteins.”
Genome sequence analysis
CRISPRFinder [69] and the CRISPR recognition tool (CRT) [70] was used to search for Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs) using the default settings. Synteny plots were created using the Nano+Bio-Center computational biology software (http://nbc3.biologie.uni-kl.de/). Comparisons were made at the DNA level using Nucmer and then the protein level using Promer by translating the DNA sequences in all six reading frames. Analyses of nucleotide content and sequence read statistics were performed using Blast+ on a 64 bit Linux server and using custom Matlab and Perl code.
Supporting Information
Phase contrast images of the five single cells subjected to single-cell whole genome amplification and genome sequencing. The small cells are indicated by circles; scale bar corresponds to 1 micrometer.
(TIF)
GC content of raw 454 reads from the 5 single cells and the AOA enrichment. The GC content of the filtered set of reads used for the final assembly of N. limnia and simulated reads from the N. maritimus genome are also shown.
(TIF)
Synteny plots relating N. limnia to the N. maritimus and C. symbiosum A genomes.
(TIF)
Genes in N. limnia that are putatively involved in ammonia oxidation and respiration (Table modified from Walker et al., 2010).
(XLS)
Movie shows last stage of sorting cell 90507.21 (occurring in the area on the device beyond the second gate valve). The laser trap is fixed near the end of the fiducial arrow. In the movie, the cell is moved some 10 s of micron before the stage is stopped. The trap beam is interrupted when the stage is stopped, freeing the cell to diffuse away. The pixels visible in the fiducial arrow are 0.106 micron wide.
(AVI)
The genome sequence is provided as a fasta file.
(TXT)
A spreadsheet containing the annotation data is provided. At the time of writing, this and the genome sequence data are in submission at the NCBI.
(XLSX)
Acknowledgments
We thank Richard White for shotgun library preparation and sequencing and Lolita Penland for sequencing the paired-end libraries. We greatly appreciate Eoghan Harrington for allowing the use of and providing support for SmashCell in beta release form (Harrington, Bioinformatics 2010, submitted), and would like to thank Jianbin Wang and Joshua Weinstein for sharing their analysis of errors arising from MDA of single template molecules.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported in part by National Science Foundation Grants MCB-0604270 and OCE-0847266 (to C.A.F.), NIH Director's Pioneer Award and NIH 5R01HG004863-02 (to S.R.Q.) and an EPA STAR Graduate Fellowship (to A.C.M.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Fuhrman JA, McCallum K, Davis AA. Novel major archaebacterial group from marine plankton. Nature. 1992;356:148–149. doi: 10.1038/356148a0. [DOI] [PubMed] [Google Scholar]
- 2.DeLong EF. Archaea in coastal marine environments. Proc Natl Acad Sci U S A. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karner MB, DeLong EF, Karl DM. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409:507–510. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- 4.Herndl GJ, Reinthaler T, Teira E, van Aken H, Veth C, et al. Contribution of Archaea to total prokaryotic production in the deep Atlantic Ocean. Appl Environ Microbiol. 2005;71:2303–2309. doi: 10.1128/AEM.71.5.2303-2309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teira E, Lebaron P, van Aken H, Herndl GJ. Distribution and activity of Bacteria and Archaea in the deep water masses of the North Atlantic. Limnology and Oceanography. 2006;51:2131–2144. [Google Scholar]
- 6.Kirchman DL, Elifantz H, Dittel AI, Malmstrom RR, Cottrell MT. Standing stocks and activity of Archaea and Bacteria in the western Arctic Ocean. Limnology and Oceanography. 2007;52:495–507. [Google Scholar]
- 7.Buckley DH, Graber JR, Schmidt TM. Phylogenetic analysis of nonthermophilic members of the kingdom Crenarchaeota and their diversity and abundance in soils. Applied and Environmental Microbiology. 1998;64:4333–4339. doi: 10.1128/aem.64.11.4333-4339.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandaa RA, Enger O, Torsvik V. Abundance and diversity of Archaea in heavy-metal-contaminated soils. Applied and Environmental Microbiology. 1999;65:3293–3297. doi: 10.1128/aem.65.8.3293-3297.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ochsenreiter T, Selezi D, Quaiser A, Bonch-Osmolovskaya L, Schleper C. Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environmental Microbiology. 2003;5:787–797. doi: 10.1046/j.1462-2920.2003.00476.x. [DOI] [PubMed] [Google Scholar]
- 10.Kemnitz D, Kolb S, Conrad R. High abundance of Crenarchaeota in a temperate acidic forest soil. Fems Microbiology Ecology. 2007;60:442–448. doi: 10.1111/j.1574-6941.2007.00310.x. [DOI] [PubMed] [Google Scholar]
- 11.Leininger S, Urich T, Schloter M, Schwark L, Qi J, et al. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature. 2006;442:806–809. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- 12.Wuchter C, Abbas B, Coolen MJ, Herfort L, van Bleijswijk J, et al. Archaeal nitrification in the ocean. Proc Natl Acad Sci U S A. 2006;103:12317–12322. doi: 10.1073/pnas.0600756103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mincer TJ, Church MJ, Taylor LT, Preston C, Kar DM, et al. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environmental Microbiology. 2007;9:1162–1175. doi: 10.1111/j.1462-2920.2007.01239.x. [DOI] [PubMed] [Google Scholar]
- 14.Herfort L, Schouten S, Abbas B, Veldhuis MJW, Coolen MJL, et al. Variations in spatial and temporal distribution of Archaea in the North Sea in relation to environmental variables. Fems Microbiology Ecology. 2007;62:242–257. doi: 10.1111/j.1574-6941.2007.00397.x. [DOI] [PubMed] [Google Scholar]
- 15.Beman JM, Popp BN, Francis CA. Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. Isme Journal. 2008;2:429–441. doi: 10.1038/ismej.2007.118. [DOI] [PubMed] [Google Scholar]
- 16.Konstantinidis KT, Braff J, Karl DM, DeLong EF. Comparative Metagenomic Analysis of a Microbial Community Residing at a Depth of 4,000 Meters at Station ALOHA in the North Pacific Subtropical Gyre. Applied and Environmental Microbiology. 2009;75:5345–5355. doi: 10.1128/AEM.00473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Church MJ, Wai B, Karl DM, DeLong EF. Abundances of crenarchaeal amoA genes and transcripts in the Pacific Ocean. Environmental Microbiology. 2010;12:679–688. doi: 10.1111/j.1462-2920.2009.02108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beman JM, Sachdeva R, Fuhrman JA. Population ecology of nitrifying Archaea and Bacteria in the Southern California Bight. Environmental Microbiology. 2010;12:1282–1292. doi: 10.1111/j.1462-2920.2010.02172.x. [DOI] [PubMed] [Google Scholar]
- 19.Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nature Reviews Microbiology. 2008;6:245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- 20.Spang A, Hatzenpichler R, Brochier-Armanet C, Rattei T, Tischler P, et al. Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol. 2010;18:331–340. doi: 10.1016/j.tim.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 21.Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, et al. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 22.Santoro AE, Francis CA, de Sieyes NR, Boehm AB. Shifts in the relative abundance of ammonia-oxidizing bacteria and archaea across physicochemical gradients in a subterranean estuary. Environ Microbiol. 2008;10:1068–1079. doi: 10.1111/j.1462-2920.2007.01547.x. [DOI] [PubMed] [Google Scholar]
- 23.Mosier A, Francis C. Relative abundance and diversity of ammonia-oxidizing archaea and bacteria in the San Francisco Bay estuary. Environmental microbiology. 2008;10:3002–3016. doi: 10.1111/j.1462-2920.2008.01764.x. [DOI] [PubMed] [Google Scholar]
- 24.de la Torre JR, Walker CB, Ingalls AE, Konneke M, Stahl DA. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing crenarchaeol. Environ Microbiol. 2008;10:810–818. doi: 10.1111/j.1462-2920.2007.01506.x. [DOI] [PubMed] [Google Scholar]
- 25.Hatzenpichler R, Lebedeva EV, Spieck E, Stoecker K, Richter A, et al. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci U S A. 2008;105:2134–2139. doi: 10.1073/pnas.0708857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marcy Y, Ouverney C, Bik EM, Losekann T, Ivanova N, et al. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Natl Acad Sci U S A. 2007;104:11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woyke T, Tighe D, Mavromatis K, Clum A, Copeland A, et al. One bacterial cell, one complete genome. Plos One. 2010;5:e10314. doi: 10.1371/journal.pone.0010314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dick GJ, Andersson AF, Baker BJ, Simmons SL, Thomas BC, et al. Community-wide analysis of microbial genome sequence signatures. Genome Biol. 2009;10:R85. doi: 10.1186/gb-2009-10-8-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moin NS, Nelson KA, Bush A, Bernhard AE. Distribution and diversity of archaeal and bacterial ammonia oxidizers in salt marsh sediments. Appl Environ Microbiol. 2009;75:7461–7468. doi: 10.1128/AEM.01001-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marcy Y, Ishoey T, Lasken RS, Stockwell TB, Walenz BP, et al. Nanoliter reactors improve multiple displacement amplification of genomes from single cells. PLoS Genet. 2007;3:1702–1708. doi: 10.1371/journal.pgen.0030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woyke T, Xie G, Copeland A, Gonzalez JM, Han C, et al. Assembling the marine metagenome, one cell at a time. Plos One. 2009;4:e5299. doi: 10.1371/journal.pone.0005299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodrigue S, Malmstrom RR, Berlin AM, Birren BW, Henn MR, et al. Whole genome amplification and de novo assembly of single bacterial cells. Plos One. 2009;4:e6864. doi: 10.1371/journal.pone.0006864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashkin A, Dziedzic JM, Bjorkholm JE, Chu S. Observation of a single-beam gradient force optical trap for dielectric particles. Opt Lett. 1986;11:288. doi: 10.1364/ol.11.000288. [DOI] [PubMed] [Google Scholar]
- 35.Neuman KC, Block SM. Optical trapping. Rev Sci Instrum. 2004;75:2787–2809. doi: 10.1063/1.1785844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White RA, 3rd, Blainey PC, Fan HC, Quake SR. Digital PCR provides sensitive and absolute calibration for high throughput sequencing. BMC Genomics. 2009;10:116. doi: 10.1186/1471-2164-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myllykallio H, Lopez P, Lopez-Garcia P, Heilig R, Saurin W, et al. Bacterial mode of replication with eukaryotic-like machinery in a hyperthermophilic archaeon. Science. 2000;288:2212–2215. doi: 10.1126/science.288.5474.2212. [DOI] [PubMed] [Google Scholar]
- 38.Richter M, Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Booth IR, Edwards MD, Black S, Schumann U, Miller S. Mechanosensitive channels in bacteria: signs of closure? Nature Reviews Microbiology. 2007;5:431–440. doi: 10.1038/nrmicro1659. [DOI] [PubMed] [Google Scholar]
- 40.Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci U S A. 2010;107:8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burg MB, Ferraris JD. Intracellular organic osmolytes: function and regulation. J Biol Chem. 2008;283:7309–7313. doi: 10.1074/jbc.R700042200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 43.Margolin W. FtsZ and the division of prokaryotic cells and organelles. Nat Rev Mol Cell Biol. 2005;6:862–871. doi: 10.1038/nrm1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindas AC, Karlsson EA, Lindgren MT, Ettema TJ, Bernander R. A unique cell division machinery in the Archaea. Proc Natl Acad Sci U S A. 2008;105:18942–18946. doi: 10.1073/pnas.0809467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in archaea. Science. 2008;322:1710–1713. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holmes VF, Cozzarelli NR. Closing the ring: links between SMC proteins and chromosome partitioning, condensation, and supercoiling. Proc Natl Acad Sci U S A. 2000;97:1322–1324. doi: 10.1073/pnas.040576797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lurz R, Grote M, Dijk J, Reinhardt R, Dobrinski B. Electron microscopic study of DNA complexes with proteins from the Archaebacterium Sulfolobus acidocaldarius. EMBO J. 1986;5:3715–3721. doi: 10.1002/j.1460-2075.1986.tb04705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo X, Schwarz-Linek U, Botting CH, Hensel R, Siebers B, et al. CC1, a novel crenarchaeal DNA binding protein. J Bacteriol. 2007;189:403–409. doi: 10.1128/JB.01246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ouverney CC, Fuhrman JA. Marine planktonic archaea take up amino acids. Appl Environ Microbiol. 2000;66:4829–4833. doi: 10.1128/aem.66.11.4829-4833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ingalls AE, Shah SR, Hansman RL, Aluwihare LI, Santos GM, et al. Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc Natl Acad Sci U S A. 2006;103:6442–6447. doi: 10.1073/pnas.0510157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Varela MM, van Aken HM, Sintes E, Herndl GJ. Latitudinal trends of Crenarchaeota and Bacteria in the meso- and bathypelagic water masses of the Eastern North Atlantic. Environ Microbiol. 2008;10:110–124. doi: 10.1111/j.1462-2920.2007.01437.x. [DOI] [PubMed] [Google Scholar]
- 52.Arp DJ, Sayavedra-Soto LA, Hommes NG. Molecular biology and biochemistry of ammonia oxidation by Nitrosomonas europaea. Arch Microbiol. 2002;178:250–255. doi: 10.1007/s00203-002-0452-0. [DOI] [PubMed] [Google Scholar]
- 53.Hooper A, Mai T, Balny C. Kinetics of reduction by substrate or dithionite and heme-heme electron transfer in the multiheme hydroxylamine oxidoreductase. European Journal of Biochemistry. 2005;141:565–571. doi: 10.1111/j.1432-1033.1984.tb08230.x. [DOI] [PubMed] [Google Scholar]
- 54.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 55.Hallam SJ, Mincer TJ, Schleper C, Preston CM, Roberts K, et al. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 2006;4:e95. doi: 10.1371/journal.pbio.0040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hallam SJ, Konstantinidis KT, Putnam N, Schleper C, Watanabe Y, et al. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc Natl Acad Sci U S A. 2006;103:18296–18301. doi: 10.1073/pnas.0608549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martens-Habbena W, Berube P, Urakawa H, de La Torre J, Stahl D. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature. 2009;461:976–979. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- 58.Burton SA, Prosser JI. Autotrophic ammonia oxidation at low pH through urea hydrolysis. Appl Environ Microbiol. 2001;67:2952–2957. doi: 10.1128/AEM.67.7.2952-2957.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci U S A. 2005;102:14683–14688. doi: 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pernthaler A, Pernthaler J. Fluorescence in situ hybridization for the identification of environmental microbes. Methods Mol Biol. 2007;353:153–164. doi: 10.1385/1-59745-229-7:153. [DOI] [PubMed] [Google Scholar]
- 61.Stahl DA, Amann RI. Development and application of nucleic acid probes in bacterial systematics. In: Stackebrandt E, Goodfellow M, editors. Sequencing and hybridization techniques in bacterial systematics. Chichester, England: John Wiley and Sons; 1991. pp. 205–248. [Google Scholar]
- 62.Amann RI, Zarda B, Stahl DA, Schleifer KH. Identification of individual prokaryotic cells by using enzyme-labeled, rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1992;58:3007–3011. doi: 10.1128/aem.58.9.3007-3011.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Daims H, Bruhl A, Amann R, Schleifer KH, Wagner M. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Systematic and Applied Microbiology. 1999;22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- 64.Maddison WP. Testing character correlation using pairwise comparisons on a phylogeny. J Theor Biol. 2000;202:195–204. doi: 10.1006/jtbi.1999.1050. [DOI] [PubMed] [Google Scholar]
- 65.DeSantis TZ, Jr, Hugenholtz P, Keller K, Brodie EL, Larsen N, et al. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34:W394–399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ludwig W, Strunk O, Westram R, Richter L, Meier H, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35:W52–57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bland C, Ramsey TL, Sabree F, Lowe M, Brown K, et al. CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics. 2007;8:209. doi: 10.1186/1471-2105-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phase contrast images of the five single cells subjected to single-cell whole genome amplification and genome sequencing. The small cells are indicated by circles; scale bar corresponds to 1 micrometer.
(TIF)
GC content of raw 454 reads from the 5 single cells and the AOA enrichment. The GC content of the filtered set of reads used for the final assembly of N. limnia and simulated reads from the N. maritimus genome are also shown.
(TIF)
Synteny plots relating N. limnia to the N. maritimus and C. symbiosum A genomes.
(TIF)
Genes in N. limnia that are putatively involved in ammonia oxidation and respiration (Table modified from Walker et al., 2010).
(XLS)
Movie shows last stage of sorting cell 90507.21 (occurring in the area on the device beyond the second gate valve). The laser trap is fixed near the end of the fiducial arrow. In the movie, the cell is moved some 10 s of micron before the stage is stopped. The trap beam is interrupted when the stage is stopped, freeing the cell to diffuse away. The pixels visible in the fiducial arrow are 0.106 micron wide.
(AVI)
The genome sequence is provided as a fasta file.
(TXT)
A spreadsheet containing the annotation data is provided. At the time of writing, this and the genome sequence data are in submission at the NCBI.
(XLSX)