Summary
The genetics of renal cancer is dominated by inactivation of the VHL tumour suppressor gene in clear cell carcinoma (ccRCC), the commonest histological subtype. A recent large-scale screen of ~3500 genes by PCR-based exon re-sequencing identified several new cancer genes in ccRCC including UTX (KDM6A)1, JARID1C (KDM5C) and SETD22. These genes encode enzymes that demethylate (UTX, JARID1C) or methylate (SETD2) key lysine residues of histone H3. Modification of the methylation state of these lysine residues of histone H3 regulates chromatin structure and is implicated in transcriptional control3. However, together these mutations are present in fewer than 15% of ccRCC, suggesting the existence of additional, currently unidentified cancer genes. Here, we have sequenced the protein coding exome in a series of primary ccRCC and report the identification of the SWI/SNF chromatin remodeling complex gene PBRM14 as a second major ccRCC cancer gene, with truncating mutations in 41% (92/227) of cases. These data further elucidate the somatic genetic architecture of ccRCC and emphasize the marked contribution of aberrant chromatin biology.
Exome sequencing based on a solution phase capture approach5 was performed on seven cases of ccRCC, three of which carry VHL mutations, and matching normal DNAs (See Supplementary information and Supplementary Table 1). Captured material was sequenced using 76 basepair paired-end reads on the Illumina GAIIx platform. After read alignment, variant calling was performed using a naïve Bayesian classifier algorithm for substitutions and a split-read mapping approach (PinDel6 with substantial cancer-aware output filtering) for insertion/deletions (See Supplementary Material for details). These algorithms aim to identify somatically acquired coding and splice-site variants (i.e. present in the tumour but not in the matching normal), and all mutations reported here were confirmed by PCR-based capillary sequencing. 156 somatic mutations were identified, of which 92 were missense, 9 nonsense, 1 canonical splice site, 1 stop codon read-through, 11 frameshift and 42 synonymous (Supplementary Table 2).
In four cases truncating mutations were indentified in PBRM1. PBRM1 maps to chromosome 3p21 and encodes the BAF180 protein, the chromatin targeting subunit of the PBAF SWI/SNF chromatin remodelling complex7. The gene is comprised of 6 bromodomains involved in binding acetylated lysine residues on histone tails, 2 bromo-adjacent homology domains important in protein-protein interaction and an HMG DNA binding domain4. PBAF complex-mediated chromatin remodelling is implicated in replication, transcription, DNA repair and control of cell proliferation/differentiation4,7. The SMARCB1 and BRG1 components of this complex have inactivating mutations in rhabdoid tumours8,9 and BRG1 mutations have been reported in multiple tumour types10. The PBRM1 mutations included three frame-shifting insertions and a nonsense mutation; all judged to be homozygous from SNP array and mutant allele read count data. PBRM1 was not included in our previous PCR-based sequencing screen2 and was the only gene, apart from VHL, with recurrent truncating mutations in the seven cases screened.
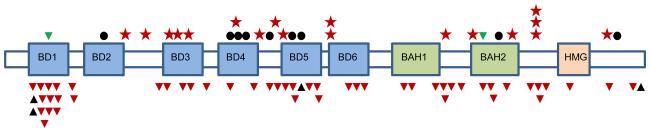
We next sequenced PBRM1 in a further 257 RCC cases, including 36 cases of papillary, chromophobe and other non-ccRCC cancers. Truncating mutations were identified in a remarkable 88/257 (34%) (Figure 1) of cases, all diagnosed as ccRCC (for full data see Supplementary Tables 3, 4). PBRM1 mutations were all found in the context of chromosome 3p loss of heterozygosity (38/38) where SNP array data was available (http://www.sanger.ac.uk/cgi-bin/genetics/CGP/cghviewer/CghHome.cgi). Two in-frame deletion mutations were identified – a predicted 6 amino-acid deletion (p.M1209_E1214delMFYKKE) in the second BAH (bromo-adjacent homology) domain likely involved in protein-protein interactions within the SWI/SNF complex4 and deletion of an isoleucine codon (Ile57) in the first bromodomain (Figure 1). Both deletions remove amino acids conserved to C elegans and both were in cases with 3p LOH. The ratio of nine missense to zero silent mutations suggests that a proportion of the missense mutations are likely to be pathogenic. Six of nine missense mutations occur in bromodomains and one in the second BAH domain (Figure 1). The bromodomains of PBRM1 have been shown to have preferential binding to different acetylated lysine configurations of histone tails, suggesting they may contribute to “reading” of the histone code11. The likelihood of the missense mutations having functional impact was assessed using a scoring system calibrated with protein domain alignments from Pfam (see Supplementary Methods). Three missense mutations (p.T232P, p.A597D and p.H1204P) could be scored with these alignments. This set of mutations was predicted to be deleterious, having a significantly lower mean score than a typical null set of in silico generated random missense mutations falling onto the scorable parts of the gene (p-value 0.01 Figure 2), making these mutations interesting candidates for functional studies.
Figure 1. PBRM1 somatic mutations.
Representation of PBRM1 transcript with boxes BR1-BR6, BAH1-2 and HMG indicating the positions of the bromodomains 1-6, bromo-adjacent homology domains and high-mobility group domain, respectively. Relative positions of mutations are indicated by symbols. Stars – nonsense, dots – missense, red triangles – frameshift deletions, black triangles – frameshift insertions and green triangles – in-frame deletions. Splice-site mutations are not depicted.
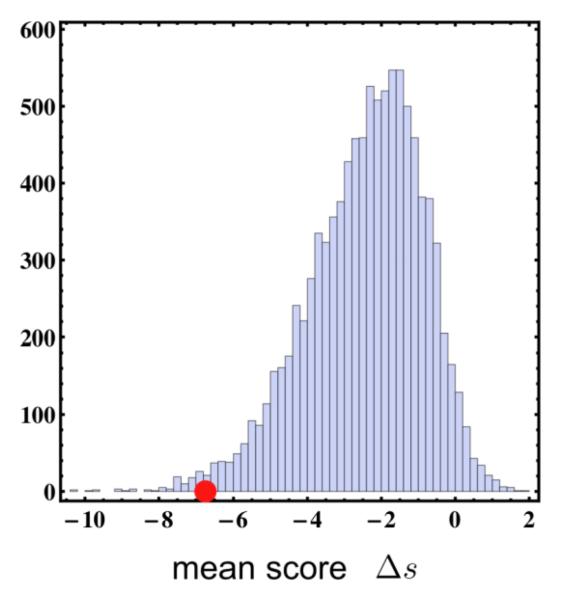
Figure 2. Analysis of PBRM1 missense mutations.
Bars represent histogram of the mean score of in silico generated random missense mutations (10,000 sets of three mutations that can be scored) and the red circle denotes the mean score of the somatic mutations that could be scored (T232P □s = −7.78, A597D □s = −9.69, H1204P □s = −2.76). The somatic set is significantly different from the null set (p-value 0.01). They have a higher negative mean score and are thus predicted to be more deleterious on average.
Four PBRM1 truncating mutations have been previously described in breast cancer12. Although there is frequent 3p21 LOH in small-cell lung cancer, no evidence for PBRM1 inactivation was found13. To further evaluate the contribution of PBRM1 mutation in human cancer, copy number was evaluated and the coding exons were sequenced through a series of 727 cancer cell lines of various histologies. SNP array copy number analysis (http://www.sanger.ac.uk/cgi-bin/genetics/CGP/cghviewer/CghHome.cgi) identified one homozygous deletion in the HCC-1143 breast cancer cell line, previously described12. Sequencing analysis identified five homozygous truncating mutations (Supplementary Table 5). Frame-shifting deletions were identified in the VHL-mutant A704 renal cancer, NCI-H2196 small-cell lung cancer and TGBC24TKB gall bladder cancer lines. Nonsense mutations were identified in the NCI-H226 squamous-cell lung cancer and PANC-10-05 pancreatic adenocarcinoma lines. Interestingly, a PBRM1 truncating mutation has been reported in a comprehensive pancreatic cancer mutational screen14.
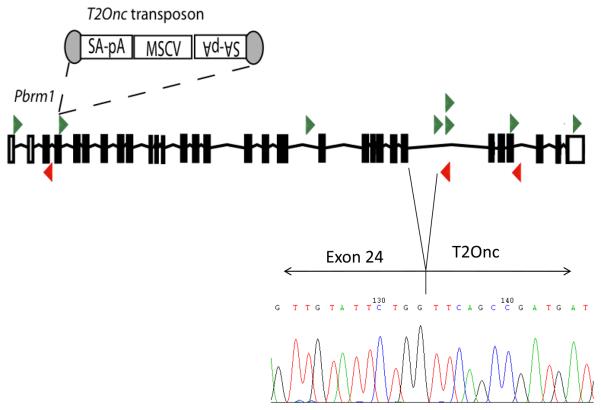
To obtain further support that PBRM1 can act as a cancer gene, we examined data from several insertional mutagenesis screens in mice. Analyses of transposon insertion sites from a forward genetic screen performed using a conditional Sleeping Beauty transposon system15 in a mouse pancreatic cancer model16 revealed a significant enrichment of insertion events in Pbrm1 amongst all genes hit using Monte Carlo simulation analyses as previously described17. Insertions were found in pancreatic dysplasia, intraductal (panIN) and high grade invasive tumours suggesting Pbrm1 inactivation is an early event in this model. The mixed forward and reverse pattern of insertions is indicative of inactivation, as demonstrated by RT-PCR showing premature termination of the Pbrm1 cDNA via splicing into the inserted transposon (Figure 3). These data suggest that loss of Pbrm1 cooperates with Kras in driving pancreatic tumour development in this model. Intriguingly, Setd2, previously implicated human ccRCC, was also found to rank significantly in frequency among all insertion sites and two tumours had both Setd2 and Pbrm1 insertions. These comparative oncogenomic data provide independent support for PBRM1 as a cancer gene and suggest further investigation of the role of PBRM1 (and SETD2) in human pancreatic cancer is warranted.
Figure 3. Pbrm1 is frequently mutated in a mouse model of pancreatic cancer.
To identify genes that co-operate with K-Ras in the formation of pancreatic cancer a conditional allele of K-RasG12D and Pdx1-Cre were combined with a conditional Sleeping Beauty transposase driver and the T2Onctg transposon donor allele29. Expression of Cre results in expression of K-RasG12D and transposon mobilization within the epithelial compartment of the pancreas. Isolation of the transposon insertion sites from a panel of 153 pancreatic cancers and pre-neoplastic lesions generated from this model revealed a common insertion site in Pbrm1 suggesting that loss of Pbrm1 co-operates with K-RasG12D in pancreatic cancer development. Statistical analysis was performed as previously described30. Transposon insertions in the forward strand of Pbrm1 are shown in green. Insertions in the reverse orientation are shown in red. A chromatogram from sequencing of RT-PCR products from one tumour is shown demonstrating splicing of exon 24 of Pbrm1 into the inserted transposon, thus truncating the transcript.
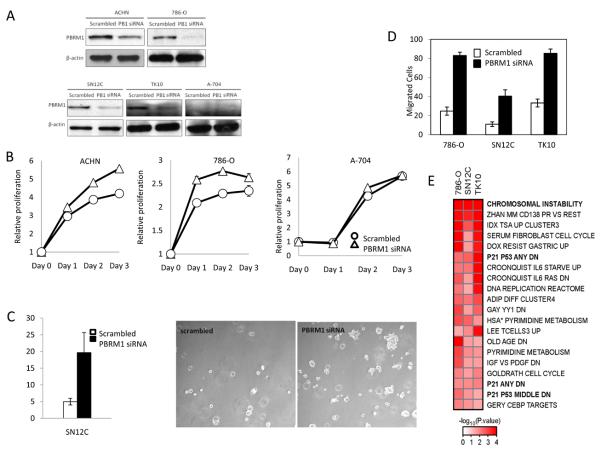
Abrogation of PBRM1 expression via siRNA knockdown in ccRCC cell lines was investigated to assess possible consequences of PBRM1 loss. Greater than 60% knockdown of PBRM1 RNA and protein resulted in a significant increase in proliferation 4/5 RCC lines (Figure 4A, B and Supplemental Information). No effect was seen, however, in A704 which carries a homozygous truncating PBRM1 mutation and expresses no PBRM1, confirming the specificity of the assay. Further, knockdown of PBRM1 resulted in significantly increased colony formation in soft-agar and increased cell migration (Figure 4C, D), indicative of an increase in transformed phenotype. Taken together, these data support PBRM1 having a tumour suppressor role in ccRCC.
Figure 4. Knockdown of PBRM1 expression in RCC cell lines.
(A) Verification of PBRM1 knockdown by western blotting. (B)Silencing PBRM1 increased the proliferation of ACHN and 786-O with wild type PBRM1, but not A704 with a homozygous PBRM1 truncating mutation. Data represent means of triplicate experiments with standard deviation, p<0.01. (C) Knockdown of PBRM1 enhanced colony formation in SN12C cells. Data represent means of triplicate experiments with standard deviation, p<0.01. (D) Knockdown of PBRM1 enhanced cell migration in 786-O, SN12C and TK10 cells. Data represent means of triplicate experiments with standard deviation, p<0.01. (E) Gene sets that are most significantly deregulated following PBRM1 knockdown in three RCC cell lines using curated gene sets obtained from MSigDB (http://www.broadinstitute.org/gsea/msigdb/) and additional curated gene sets obtained from the PGSEA package (see Supplemental Material for details).
Transcriptional profiling before and after PBRM1 knockdown was performed using gene expression microarrays. Gene set enrichment analysis following PBRM1 knockdown showed that PBRM1 activity regulates pathways associated with chromosomal instability and cellular proliferation (Figure 4E, Supplementary Table 6), the latter being consistent with previous studies identifying PBRM1 as critical transcriptional regulator of p21/CDKN1A in breast cancer cell lines12 and work showing that PBRM1/Baf180 is implicated in regulating TP53 mediated replicative senescence 18. The PBAF complex has been shown to localise at kinetochores during mitosis19 and SMARCB1 has been implicated in spindle checkpoint control20, which would support the loss of PBRM1 giving rise to a chromosomal instability/spindle checkpoint expression phenotype. It may be of interest to further explore spindle checkpoint control in PBRM1 mutated ccRCC as a potential therapeutic opportunity.
Previous work has demonstrated that VHL loss alone is insufficient for ccRCC tumourigenesis arguing the need for additional genetic events21,22 (Teh, unpublished) and has further suggested the existence of a 3p21 “gatekeeper” ccRCC mutation based on LOH studies23. The data presented here strongly suggest that inactivation of PBRM1 comprises this second major mutation in ccRCC development. Nearly all (36/38) PBRM1 mutant cases fall into the hypoxia signature group as described previously2, including 13/14 cases without demonstrable VHL point mutations where expression data is available – further indicating the importance of PBRM1 in typical ccRCC development. The SWI/SNF complex has been implicated in the normal cellular response to hypoxia, with impairment of the complex rendering cells resistant to hypoxia-induced cell cycle arrest24, which would be consistent with selection for frequent loss of PBRM1 in ccRCC. Multiple cancers have apparently concomitant VHL, PBRM1 and SETD2 mutations, with all three genes mapping to chromosome 3p, suggesting that the mutations are non-redundant functionally. Half (55/107) of cases in this series with a demonstrable VHL mutation 2 have a PBRM1 mutation. Strikingly, all 9 cases with a SETD2 mutation have a mutation in either PBRM1 or VHL, with 6 of 9 cases having mutations in all three genes. Physical linkage of these three ccRCC cancer genes together with their potential interaction may be the key driver for the large scale 3p LOH seen in most cases of ccRCC – being particularly parsimonious in requiring only four genetic events to unmask three tumour suppressor genes as opposed to six if the genes were on different chromosomes.
Several other mutated genes of potential interest were identified. In particular, ARID1A encoding the BAF250A subunit of the SWI/SNF complex was found to have two heterozygous missense mutations - p.R1020K,c.3059G>A and p.L1872P,c.5615T>C. Both cases (PD2126, PD2127) have a PBRM1 truncating mutation. Two homozygous ARID1A deletions were found in SNP 6.0 data (http://www.sanger.ac.uk/cgi-bin/genetics/CGP/conan/search.cgi) in the LB1047-RCC ccRCC and NCI-SNU-5 gastric carcinoma cell lines and loss of ARID1A expression has been reported in RCC25. Frequent truncating ARID1A truncating mutations have recently been reported in clear cell ovarian carcinoma 26,27. These data all point to ARID1A being a cancer gene, likely operative in ccRCC. PD2127 was also found to have a heterozygous truncating mutation in ARID5B, related to ARID1A and recently implicated in childhood acute lymphoblastic leukaemia susceptibility28. The extent to which the other mutated genes identified here contribute to ccRCC will await large-scale follow-up screens. Similarly, exome and whole genome sequencing on a large number of cases is likely to yield further insights.
The identification of a second major cancer gene in ccRCC further defines the genetic and molecular architecture of this tumour type. It is remarkable that PBRM1, like the majority of the other non-VHL mutated cancer genes identified in ccRCC, is involved in chromatin regulation – again at least in part at the level of histone H3 modification and recognition. Understanding the contribution of PBRM1 mutation to clinical disease progression and outcome as well the potential for exploiting SWI/SNF complex abrogation therapeutically are important future areas of renal cancer research.
Methods Summary
DNA samples from ccRCC patients tumour and matching normal were all obtained under local IRB and LREC approvals for this study and processed as previously described2. DNA fragmentation, library preparation and solution phase hybrid capture were according to manufacturer instructions (Agilent Technologies, US) and modified from previously published protocols5. Capillary-based Sanger sequencing for confirmations and PBRM1 followup were done as previously described2 with manual inspection of all sequencing traces. mRNA was extracted from snap-frozen mouse pancreatic lesions and subjected to RT-PCR using a nested PCR approach utilising primers of mouse Pbrm1 exon 23/24 and the Carp-β-Actin Splice acceptor sequence of the T2Onc transposon cassette. Resulting bands were gel-purified and subjected to capillary-based Sanger sequencing. PBRM1 or scrambled control siRNAs (Santa Cruz, CA) were transfected into ccRCC cell lines using Lipofectamine 2000 (Invitrogen, CA) according to the manufacturer's conditions. Real-time PCR and western blotting were all done utilising standard protocols essentially as described1. Expression analyses were carried out as previously described2.
Supplementary Material
Acknowledgements
PAF and MRS would like to acknowledge the Wellcome Trust for support under grant reference 077012/Z/05/Z and Allyson Coffey, Dan Turner and Lyra Mamanova for assistance with the exon capture. KF, KD and BTT acknowledge the support of the Van Andel Research Institute. BTT would like to acknowledge support from the Lee Foundation. IV is supported by a fellowship from The International Human Frontier Science Program Organization. DJA acknowledges the support of Cancer Research UK. DT and PP-M acknowledge the support of the University of Cambridge, Cancer Research UK and Hutchison Whampo and thank Dr. Will Howatt, Mr. Allen Hazelhurst and colleagues in the CRI core facilities for their support. BTT would like to dedicate this work to Tat Hock Teh.
Footnotes
Author information
Exome sequence data have been deposited at the European Genome-Phenome Archive (http://www.ebi.ac.uk/ega/) hosted by the European Bioinformatics Institute under accession EGAS00001000006 and expression data has been deposited with Gene Expresson Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession GEO22316.
The authors declare no competing financial interests.
References
- 1.van Haaften G, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–3. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dalgliesh GL, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kouzarides T. Chromatin Modifications and Their Function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Thompson M. Polybromo-1: The chromatin targeting subunit of the PBAF complex. Biochimie. 2009;91:309–319. doi: 10.1016/j.biochi.2008.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gnirke A, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotech. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–1668. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- 8.Schneppenheim R, et al. Germline Nonsense Mutation and Somatic Inactivation of SMARCA4/BRG1 in a Family with Rhabdoid Tumor Predisposition Syndrome. The American Journal of Human Genetics. 2010;86:279–284. doi: 10.1016/j.ajhg.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Versteege I, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 10.Wong AKC, et al. BRG1, a Component of the SWI-SNF Complex, Is Mutated in Multiple Human Tumor Cell Lines. Cancer Res. 2000;60:6171–6177. [PubMed] [Google Scholar]
- 11.Chandrasekaran R, Thompson M. Polybromo-1-bromodomains bind histone H3 at specific acetyl-lysine positions. Biochemical and Biophysical Research Communications. 2007;355:661–666. doi: 10.1016/j.bbrc.2007.01.193. [DOI] [PubMed] [Google Scholar]
- 12.Xia W, et al. BAF180 Is a Critical Regulator of p21 Induction and a Tumor Suppressor Mutated in Breast Cancer. Cancer Research. 2008;68:1667–1674. doi: 10.1158/0008-5472.CAN-07-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sekine I, et al. The 3p21 candidate tumor suppressor gene BAF180 is normally expressed in human lung cancer. 2005;24:2735–2738. doi: 10.1038/sj.onc.1207694. [DOI] [PubMed] [Google Scholar]
- 14.Jones S, et al. Core Signaling Pathways in Human Pancreatic Cancers Revealed by Global Genomic Analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keng VW, et al. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat Biotech. 2009;27:264–274. doi: 10.1038/nbt.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hingorani SR, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 17.Starr TK, et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323:1747–50. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burrows AE, Smogorzewska A, Elledge SJ. Polybromo-associated BRG1-associated factor components BRD7 and BAF180 are critical regulators of p53 required for induction of replicative senescence. Proc Natl Acad Sci U S A. 2010;107:14280–14285. doi: 10.1073/pnas.1009559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xue Y, et al. The human SWI/SNF-B chromatin-remodeling complex is related to yeast Rsc and localizes at kinetochores of mitotic chromosomes. Proc Natl Acad Sci U S A. 2000;97:13015–13020. doi: 10.1073/pnas.240208597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vries RGJ, et al. Cancer-associated mutations in chromatin remodeler hSNF5 promote chromosomal instability by compromising the mitotic checkpoint. Genes Dev. 2005;19:665–670. doi: 10.1101/gad.335805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mandriota SJ, et al. HIF activation identifies early lesions in VHL kidneys: Evidence for site-specific tumor suppressor function in the nephron. Cancer Cell. 2002;1:459–468. doi: 10.1016/s1535-6108(02)00071-5. [DOI] [PubMed] [Google Scholar]
- 22.Young AP, et al. VHL loss actuates a HIF-independent senescence programme mediated by Rb and p400. Nat Cell Biol. 2008;10:361–369. doi: 10.1038/ncb1699. [DOI] [PubMed] [Google Scholar]
- 23.Steven CC, Amanda HP, Nabeel AA, Charles HCMB, Eamonn RM. Inactivation of the von Hippel-Lindau (VHL) tumour suppressor gene and allelic losses at chromosome arm 3p in primary renal cell carcinoma: Evidence for a VHL-independent pathway in clear cell renal tumourigenesis. Genes, Chromosomes and Cancer. 1998;22:200–209. doi: 10.1002/(sici)1098-2264(199807)22:3<200::aid-gcc5>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 24.Kenneth NS, Mudie S, van Uden P, Rocha S. SWI/SNF Regulates the Cellular Response to Hypoxia. Journal of Biological Chemistry. 2009;284:4123–4131. doi: 10.1074/jbc.M808491200. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, et al. Expression of p270 (ARID1A), a component of human SWI/SNF complexes, in human tumors. Int J Cancer. 2004;112:636–642. doi: 10.1002/ijc.20450. [DOI] [PubMed] [Google Scholar]
- 26.Jones S, et al. Frequent Mutations of Chromatin Remodeling Gene ARID1A in Ovarian Clear Cell Carcinoma. Science. 2010 doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiegand KC, et al. ARID1A Mutations in Endometriosis-Associated Ovarian Carcinomas. N Engl J Med. 2010 doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papaemmanuil E, et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat Genet. 2009;41:1006–1010. doi: 10.1038/ng.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- 30.Uren AG, et al. Large-Scale Mutagenesis in p19ARF- and p53-Deficient Mice Identifies Cancer Genes and Their Collaborative Networks. Cell. 2008;133:727–741. doi: 10.1016/j.cell.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.