Abstract
The human gut microbiome is a complex ecosystem composed mainly of uncultured bacteria. It plays an essential role in the catabolism of dietary fibers, the part of plant material in our diet that is not metabolized in the upper digestive tract, because the human genome does not encode adequate carbohydrate active enzymes (CAZymes). We describe a multi-step functionally based approach to guide the in-depth pyrosequencing of specific regions of the human gut metagenome encoding the CAZymes involved in dietary fiber breakdown. High-throughput functional screens were first applied to a library covering 5.4 × 109 bp of metagenomic DNA, allowing the isolation of 310 clones showing beta-glucanase, hemicellulase, galactanase, amylase, or pectinase activities. Based on the results of refined secondary screens, sequencing efforts were reduced to 0.84 Mb of nonredundant metagenomic DNA, corresponding to 26 clones that were particularly efficient for the degradation of raw plant polysaccharides. Seventy-three CAZymes from 35 different families were discovered. This corresponds to a fivefold target-gene enrichment compared to random sequencing of the human gut metagenome. Thirty-three of these CAZy encoding genes are highly homologous to prevalent genes found in the gut microbiome of at least 20 individuals for whose metagenomic data are available. Moreover, 18 multigenic clusters encoding complementary enzyme activities for plant cell wall degradation were also identified. Gene taxonomic assignment is consistent with horizontal gene transfer events in dominant gut species and provides new insights into the human gut functional trophic chain.
The human intestinal microbiome is the dense and complex ecosystem that resides in the distal part of our digestive tract. Its role in metabolizing dietary constituents (Sonnenburg et al. 2005; Flint et al. 2008; Ley et al. 2008) and in protecting the host against pathogens (Rakoff-Nahoum et al. 2004) is crucial to human health (Macdonald and Monteleone 2005; McGarr et al. 2005; Manichanh et al. 2006; Turnbaugh and Gordon 2009). It is mainly composed of commensal bacteria from the Bacteroidetes, Firmicutes, Proteobacteria, and Actinobacteria phyla (five), and of several archaeal and eukaryotic species. With up to 1012 cells per gram of feces, the bacterial abundance is estimated to reach 1000 operational taxonomic units (OTUs) per individual, 70% to 80% of the most dominant ones being subject-specific (Zoetendal et al. 1998; Tap et al. 2009). However, only 20% of the bacterial species have been successfully cultured so far (Eckburg et al. 2005). Large-scale analyses of genomic and metagenomic sequences have provided gene catalogs and statistical evidence on protein families involved in the predominant functions of the human gut microbiome (Gill et al. 2006; Kurokawa et al. 2007; Flint et al. 2008; Turnbaugh et al. 2009; Qin et al. 2010), among which the catabolism of dietary fibers is of particular interest in human nutrition and health. Dietary fibers are the components of vegetables, cereals, leguminous seeds, and fruits that are not digested in the stomach or in the small intestine, but are fermented in the colon by the gut microbiome and/or excreted in feces (Grabitske and Slavin 2008). Chemically, dietary fibers are mainly composed of complex plant cell wall polysaccharides and their associated lignin (Selvendran 1984), along with storage polysaccharides such as fructans and resistant starch (Institute of Medicine 2005). Dietary fibers have been identified as a strong positive dietary factor in the prevention of obesity, diabetes, and cardiovascular diseases (World Health Organization 2003). Because of the wide structural diversity of dietary fibers, the human gut bacteria produce a huge panel of carbohydrate active enzymes (CAZymes), with widely different substrate specificities, to degrade these compounds into metabolizable monosaccharides and disaccharides. The functions and the evolutionary relationships of CAZyme-encoding genes of the human gut microbiome are being extensively studied through functional and structural genomics investigations (Flint et al. 2008; Lozupone et al. 2008; Mahowald et al. 2009; Martens et al. 2009), which are nevertheless restricted to cultivated bacterial species. CAZyme diversity has also been described in three metagenomics studies focused on this microbiome (Gill et al. 2006; Turnbaugh et al. 2009, 2010), and these revealed the presence of at least 81 families of glycoside-hydrolases, making the human gut metagenome one of the richest source of CAZymes (Li et al. 2009). However, the proof of function of annotated genes issued from metagenomes still constitutes a goal for enzyme discovery. This can be addressed by functional screening of metagenomic libraries, in order to retrieve genes of interest. Numerous studies have provided conclusive evidence on the potential of such an approach for the identification of novel glycoside-hydrolases from various ecosystems such as soil (Rondon et al. 2000; Richardson et al. 2002; Voget et al. 2003; Pang et al. 2009), lakes (Rees et al. 2003), hot springs (Tang et al. 2006, 2008), rumen (Ferrer et al. 2005; Guo et al. 2008; Liu et al. 2008; Duan et al. 2009), rabbit (Feng et al. 2007), and insect guts (Brennan et al. 2004; for review, see Ferrer et al. 2009; Li et al. 2009; Simon and Daniel 2009; Uchiyama and Miyazaki 2009). In all cases, the identification of the gene responsible for the screened activity was carried out by sequencing only a few kilobases of metagenomic DNA. Collectively these studies have established an experimental proof of function for 35 glycoside hydrolases (from eight families) issued from metagenomes (data from the CAZy database; http://www.cazy.org/), a number that is very small considering the known CAZy diversity. Here, we examined the potential of high-throughput functional screening of large insert libraries to guide in-depth pyrosequencing of specific regions of the human gut metagenome that encode the enzymatic machinery involved in dietary fiber catabolism.
Results and Discussion
Function-based strategy to target novel CAZymes
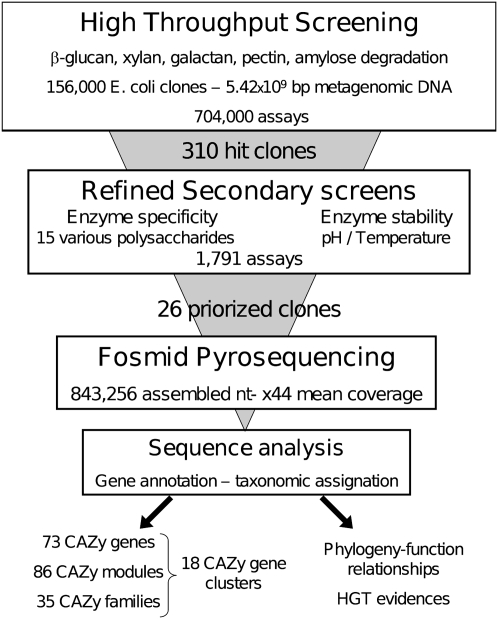
The overall strategy (Fig. 1) relies on the screening of a large metagenomic library issued from the feces of a healthy volunteer adult individual who followed a fiber-rich diet, to easily isolate genes encoding enzymes that were able to break down raw and mostly insoluble plant polysaccharides. First, the library was screened at a throughput of 200,000 clones assayed per week and per activity, using both commercial and home-made polysaccharides (Supplemental Table S1). In the secondary step, all positive clones were screened again using a panel of 15 raw and chemically modified polysaccharides of various structures (Supplemental Table S1), to distinguish different enzyme specificities toward glycosidic linkages within clones that were able to degrade the same polysaccharide in the primary screens. In parallel, enzyme pH dependency and thermostability were assayed. Then, in-depth pyrosequencing of the metagenomic DNA insert from the most interesting clones was carried out. To identify the enzymes responsible for plant polysaccharide breakdown and their microbial origin, sequence analysis was focused on taxonomic annotation of the DNA inserts and CAZyme-encoding gene annotation.
Figure 1.
Overall strategy based on the use of multi-step functional screens for gene discovery from metagenomic sequences.
Multi-step functional screening
The initial library consisted of 156,000 Escherichia coli fosmid clones, covering in total 5.46 × 109 bp of metagenomic DNA, each clone comprising a 30–40-kb DNA insert. The library was screened for the ability to hydrolyze five different polysaccharides, namely, beta-glucan, xylan, beta-(1-4)-galactan, pectin, and amylose. In total, 704,000 tests were performed, and 310 positive clones were obtained. Hit frequency varied from 0.05% to 0.8% (Supplemental Table S1). No clone degraded more than one of the substrates included in the primary screens. Secondary screening results allowed the clustering of the 310 positive clones on the basis of their ability to break down various polysaccharide structures (Supplemental Table S2). One-hundred-and-forty-two clones were able to degrade only the polysaccharide used in the primary screen, while the others could also cleave polysaccharides carrying modifications in the main chain and in the various side chains. Besides, the enzymes’ ability to work at extreme pH and high temperature was investigated for their potential use in industrial process. Enzyme stability is related to tight protein structural features, and not only to the thermotolerance of the organism they are issued from. Here, eight of the 310 positive clones maintained enzyme activity at pH 4 and/or 9, and three were still active after a 55°C heat shock. Even issued from an ecosystem regulated at 37°C, a total of 26 clones were selected from the two screening steps either for their efficiency of degradation of particularly resistant substrates, like native heteroxylans, beta-glucans, or resistant starches, and/or for their stability at various pH values or high temperatures. The percentage of clones being sequenced was thus not related to hit frequency.
Pyrosequencing and gene prediction
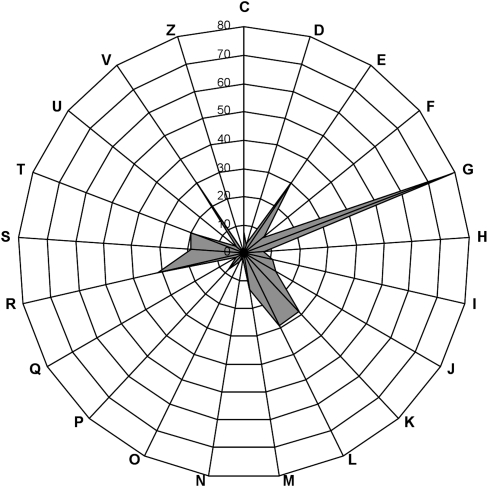
The third step of our work consisted in pyrosequencing the inserts from the 26 selected positive clones. Read assembly resulted in 27 large contigs obtained with a mean coverage sequencing depth of 44×. Two large contigs were found for clone 4. Surprisingly, three cases of partial sequence redundancy occurred for beta-glucanase, xylanase, and galactanase active clones, respectively. Excluding the vector sequences, these 27 large contigs, sizing between 8.3 and 43.8 kb, included 843,256 nt of nonredundant metagenomic DNA. The high sequencing depth allowed accurate gene prediction, gene organization, and taxonomic assignment. The total number of predicted genes sizing at least 60 nt was 665 (622 complete genes). Among the 622 complete protein sequences reported here, 349 were assigned to clusters of orthologous groups of proteins (COGs). The distribution pattern of COG-assigned proteins (Fig. 2; Supplemental Table S4) highlights the dominance of the G cluster, corresponding to proteins predicted to be involved in carbohydrate transport and metabolism. The G cluster size was found to contain 23% of COG-assigned proteins, which is drastically higher than what was previously obtained from random sequencing of the human gut metagenome (Kurokawa et al. 2007; Turnbaugh et al. 2009; Qin et al. 2010). This demonstrates the power of the functional screening steps to isolate large metagenomic DNA fragments that are enriched in genes encoding the enzymatic machinery for dietary fiber digestion.
Figure 2.
Distribution pattern of COG-assigned proteins. The genes not assignable to any COGs are not shown in this figure. (C) Energy production and conversion. (D) Cell cycle control, mitosis, and meiosis. (E) Amino acid transport and metabolism. (F) Nucleotide transport and metabolism. (G) Carbohydrate transport and metabolism. (H) Coenzyme transport and metabolism. (I) Lipid transport and metabolism. (J) Translation. (K) Transcription. (L) Replication, recombination, and repair. (M) Cell wall/membrane biogenesis. (N) Cell motility. (O) Post-translational modification, protein turnover, chaperones. (P) Inorganic ion transport and metabolism. (Q) Secondary metabolite biosynthesis, transport, and catabolism. (R) General function prediction only. (S) Function unknown. (T) Signal transduction mechanisms. (U) Intracellular trafficking and secretion. (V) Defense mechanisms. (Z) Cytoskeleton.
Taxonomic assignment of metagenomic DNA
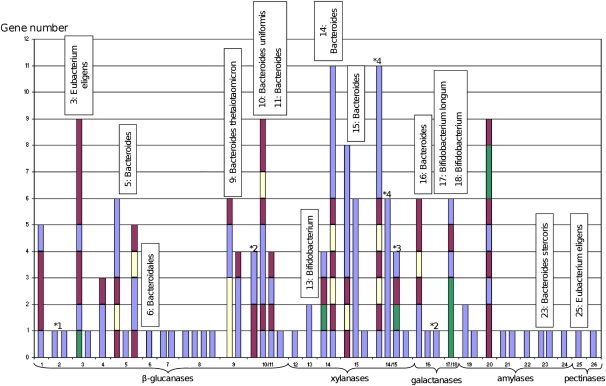
To obtain new insights into the relationships existing between bacteria taxonomy and their role in fiber metabolization, the bacterial origin of the metagenomic DNA inserts was predicted on the basis of sequence homology with the protein sequences contained in the nonredundant (NR) protein sequence database of the NCBI. The amount of assignable and unassignable metagenomic DNA fragments is biased by the number of bacterial genome sequences present in the NR database, and it is related to the highly stringent criteria (Kurokawa et al. 2007) that we used to avoid false taxonomic assignment. For all clones, the metagenomic sequences contained some genes encoding proteins without any high sequence identity with any known proteins (Supplemental Fig. S1). We thus conclude that they originate from microorganisms whose genome sequence is not (or not yet) available. Moreover, using the chosen criteria, 13 large contigs were nonassignable, one was assigned to a bacterial order, seven were assigned to one bacterial genus, and six at a bacterial species level (Fig. 3). Among them, nine corresponded to bacteria from the Bacteroidetes phylum and five to Gram-positive bacteria. This indicates that a significant number of genes originating from these bacteria were successfully expressed and produced functional proteins, even if some expression bias probably occurred by using E. coli as the recombinant host for functional screening (Gabor et al. 2004; Chen et al. 2007). Indeed, it appears that some genes that were correctly expressed in E. coli (based on the transposon mutagenesis results) were located up to 30 kb from any possible upstream vector-borne promoters. These genes came, among others, from contigs assigned to Bacteroides (i.e., prot ID ADD61481, clone 14, 30 kb; ADD61507, clone 16, 14 kb) and the Gram-positive Eubacterium (ADD61840, clone 3, 20 kb) (Supplemental Table S3). In the E. coli host, transcription of these genes was probably initiated from the native Bacteroides and Eubacterium promoters.
Figure 3.
CAZy gene clusters for each clone sequence from 1 to 26. Below the clone number is the activity for which each clone has been screened. (Blue) CAZy-encoding genes; (yellow) SusD homolog–encoding genes; (green) transport system protein–encoding genes; (purple) other genes. 14/15 shows the CAZy gene clusters of assembled sequences from these clones. Clones 10 and 11 and clones 17 and 18 have the same CAZy gene clusters; these sequences are not assembled together. On top of each bar is the taxonomic assignation of the clone when assignable, other clones are nonassigned. (*) Synteny with Roseburia intestinalis L1-82 (1); Bacteroides uniformis ATCC 8492 (2); Bacteroides stercoris ATCC 43183 (3); Bacteroides eggerthii DSM 20697 (4).
Additionally, we compared the taxonomic assignment of contigs with that of the total metagenomic DNA used for constructing the library (based on 4530 16S rDNA gene sequences) (Supplemental Fig. S2). The total bacterial diversity of the originating sample, estimated by Chao index on 16S rDNA library data sets (Supplemental Fig. S3), is consistent with the average diversity in fecal samples from healthy individuals, cumulatively reaching 9940 OTUs for 17 individuals (Tap et al. 2009). In the initial sample, the most abundant 16S rDNA sequences were assigned to five OTUs: two Eubacterium rectale (1207 sequences), Ruminococcus sp. (710 sequences), Bacteroides sp. (367 sequences), and Ruminococcus bromii (125 sequences). Surprisingly, none of the bacterial species assigned to the contigs corresponded to these five OTUs. In addition, based on 16S rDNA sequencing, some of the metagenomic fragments originated from species representing <1% of the initial sample: One 16S rDNA sequence only corresponded to Bacteroides stercoris, Bacteroides thetaiotaomicron, and Bacteroides uniformis, while 29 16S rDNA sequences corresponded to Bifidobacterium longum. Even if some cloning (Temperton et al. 2009) and expression (Gabor et al. 2004; Chen et al. 2007) biases may have occurred, and considering only taxonomic assignment to the genus level, it can be concluded that the present functionally guided strategy allows the isolation of DNA fragments from bacteria representing only a few percent of the dominant gut bacteria (like Bifidobacteria), provided that one is capable of exploring a sufficiently large sequence space.
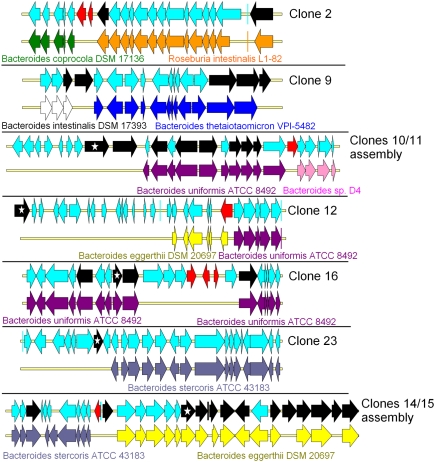
Because the frequent occurrence of horizontal gene transfer (HGT) is thought to help gut bacteria to share their advantages when facing common challenges (Roberts et al. 2008), taxonomic assignment based on sequence identity may be inconsistent with that based on 16S rDNA. It has been shown previously that the human gut metagenome is rich in conjugative transposons, integrases, and recombinases (Jones and Marchesi 2007; Kurokawa et al. 2007; Qu et al. 2008). Based on the data available in 2008, Tamames and Moya (2008) predicted that 1%–2.5% of contigs of the human gut metagenome contain probable HGT events. Moreover, the analysis of 36 bacterial gut genomes revealed that CAZyme convergence was largely due to HGT (Lozupone et al. 2008). Here, based on the analysis of only 0.84 Mb of nonredundant metagenomic sequences, we identified 11 genes predicted to encode transposases, recombinases, and integrases, assigned to COG families 3385, 4584, 5433, 3464, 3547, 4973, and 4974 (COG category L) (Supplemental Table S4). Moreover, in five cases, we observed a drastic change of DNA taxonomic assignation based on sequence homology around the gene encoding transposase, integrase, or recombinase (Fig. 4). In the case of clones 2, 11, 12, and 14/15, the first part of the contigs presented a perfect synteny with a genomic fragment from one gut bacterium, while the second part showed synteny with a fragment of a different gut bacterial genome. In the case of clone 16, the synteny with the B. uniformis ATCC 8492 genome is lost for seven genes in the middle of the contig that are not even highly similar to any B. uniformis ATCC 8492 gene. We thus hypothesize that, as for the other clones mentioned in Figure 4, such a gene organization results from gene transfers between bacterial species. For these clone sequences, the genomic heterogeneity was also confirmed by tetranucleotide frequency analysis (Supplemental Fig. S4). This provides conclusive evidence of human gut metagenome plasticity. Such a demonstration was rendered possible by the in-depth sequencing of large metagenomic DNA fragments, which provided both reliable information about gene organization and the proof that the contigable sequences originated from a single bacterial genome.
Figure 4.
Evidence of horizontal gene transfers (HGTs) in human gut metagenomic sequences. HGTs were identified when rupture was observed in gene synteny between the genes present in the metagenomic DNA fragments and their best BLASTP hits issued from sequenced genomes. For each clone, the first line represents the clone metagenomic sequence, and the second line represents the genome part in synteny with it. Each arrow represents a gene. (Red arrows) Genes encoding putative transposases or integrases; (black arrows) CAZy-encoding genes; (stars within black arrows) genes encoding the CAZymes involved in the activity detected in the primary screens, as proven by transposon insertion in the fosmid inserts.
Identification and organization of CAZyme-encoding genes
The detection of genes encoding CAZymes, which are responsible for polysaccharide degradation, was the last step of the strategy (Fig. 1). A BLAST-based sequence comparison against the CAZy database identified 73 CAZyme proteins, encoded by 65 full-length and eight truncated genes (SI). Several proteins were multimodular, resulting in a total of 86 modules assigned to 35 known CAZy families (Supplemental Table S3), corresponding mainly to polysaccharide degrading activities, including 20 glycoside-hydrolase (GH), seven carbohydrate-esterase (CE), and one polysaccharide lyase (PL) families. In order to identify the gene that is responsible for the detected activity in the primary screens, we have performed a transposon mutagenesis of the fosmid inserts. All of the proteins (labeled in Supplemental Table S3) for which an experimental proof of function is provided, were identified as CAZymes by using sequence-based analysis. They all contain a catalytic module belonging to a known GH or CE family, of which the activity described in the CAZy database is in agreement with the activity we screened for. We did not obtain any inactivated clones by transposon mutagenesis of clones 1, 5, 8, and 9. This indicates that several enzymes encoded by these fosmids may be involved in the detected activity.
Besides, many CAZymes involved in the breakdown of plant polysaccharides display a modular structure in which the catalytic domain carries one or several ancillary domains that can be catalytic, carbohydrate-binding, or of as-yet-unknown function. Four known families of carbohydrate-binding modules (CBM) and one fibronectin (FN) module were also found to be associated with catalytic modules, presumably for the attachment of enzymes to their substrates. Moreover, five of the 73 identified CAZymes (marked in Supplemental Table S3) harbored additional modules with no similarity to any known CAZy family. These families of modules of unknown function potentially represent five novel CAZy families. The precise function of these novel protein modules will be investigated by rational truncation of the corresponding proteins, in order to identify the catalytic or carbohydrate-binding function of the modules in question.
Among the 622 complete nonredundant genes, 19% were predicted to encode a signal peptide. This number increased to 38% when considering only the CAZyme-encoding genes. This is consistent with the role of these enzymes in vivo in the digestion of polysaccharide substrates that are impossible to internalize by bacterial cells. It is probable that most of the CAZymes were not secreted by E. coli cells used here as the recombinant host. Instead, CAZyme access to the insoluble polysaccharides of the functional screens was most likely due to the release of cytoplasmic proteins by E. coli cell lysis.
As demonstrated by the G COG-cluster enrichment, the present function-based strategy was very powerful in focusing the sequencing only on metagenomic DNA fragments rich in CAZyme modules. One module was found every 10 kb, with a fivefold higher frequency than that observed from random sequencing (Turnbaugh et al. 2009). The enrichment in catabolic genes can also be estimated by the glycoside hydrolase/glycosyltransferase (GH/GT) ratio. The functional screen strategy that we used led to a GH/GT ratio of 33, much higher than the 1.5 ratio obtained in the analysis of complete genomes from gut bacteria (Lozupone et al. 2008) or even the 3.4 ratio within metagenomics short reads (Turnbaugh et al. 2009). Our strategy for target-gene enrichment in metagenomes is even more efficient that those based on DNA isolation from enrichment cultures grown on polysaccharides (Grant et al. 2004) or on labeling DNA through stable isotope probing (Kalyuzhnaya et al. 2008).
The study of the organization of CAZyme-encoding genes identified here is of particular interest. Among the 73 CAZyme-encoding genes, 48 were found to constitute 18 multigenic clusters, possibly representing operon-like systems including other genes involved in carbohydrate transport and/or binding like SusD homologs and putative proteins from the TonB-dependant receptor family (Fig. 3; Martens et al. 2009). In five cases, a striking synteny was obtained with similar gene clusters from genomes of gastrointestinal tract bacteria, for which the biochemical proof of function has never been described to our knowledge. For the first time using a screening-based metagenomics approach, we describe CAZyme gene clusters involved in dietary fiber catabolism by the human gut microbiome.
Interestingly, the distribution of CAZyme gene clusters and the number of CAZyme modules and families were highly variable among the clones and found to depend on their activities. Indeed, metagenomic DNA inserts from clones able to degrade starch, contained only one to three CAZyme modules corresponding mainly to family GH13. In comparison, the DNA fragments inserted in clones able to degrade beta-glucans and xylan contained up to 17 CAZyme modules corresponding to 13 different CAZyme families. All the functions of these CAZyme modules (cellulases, hemicellulases, carbohydrate-esterases, and associated carbohydrate-binding modules) are required in vivo for the complete degradation of plant cell wall polysaccharides, whose structures are much more complex than that of starch. These operon-like clusters probably reflect the adaptation of the genetic potential of gut bacteria to the degradation of highly complex polysaccharide structures.
Finally, in order to assess how prevalent the genes we identified are among the gut microbiomes worldwide, we compared our data to the metagenome sequences currently available, issued from 124 European (Qin et al. 2010), 13 Japanese (Kurokawa et al. 2007), and 46 U.S. individuals (Gill et al. 2006; Turnbaugh et al. 2009, 2010). None of the genes we identified in our contigs was found in the U.S. and Japanese individual data sets. This was probably because we used highly stringent criteria for searching similarities with our full-length protein sequences (E-value = 0; identity ≥ 90%), in order to avoid any overestimation of the gene prevalence. In contrast, when comparing our data to the 3.3-million-gene catalog obtained from the European individuals, we identified 154 highly prevalent genes, detected in 20 individuals or more (identity ≥ 90%) (Supplemental Table S4). Among them, 33 encode CAZymes. In addition, among the 65 complete CAZyme-encoding genes of the present study, 32 matched with 100% identity to genes present in at least one individual, and six in at least 12 individuals (protein ID ADD61840, clone 3; ADD62008, clone 10; ADD62010, clone 10; ADD62011, clone 10; ADD61504, clone 16; ADD61689, clone 22) (Supplemental Table S3). These six CAZymes were found in the gut microbiomes of individuals with very distinct body mass index (lean, overweight, obese), and with different clinical status (healthy, inflammatory bowel–diseased patients). Moreover, in many cases (for clones 3, 4, 9, 10/11, 14/15, 16, 19, 20, 26), the genes surrounding these highly prevalent CAZymes were also present in several individuals. These results show the power of such an activity-based functional metagenomics approach, even when applied on a single sample, to provide an experimental proof of function to highly prevalent genes and gene clusters of the human gut microbiome. This also underlines the interest of coupling sequence-based and activity-based metagenomics to investigate the gut microbiota functions and to measure the prevalence and abundance of targeted genes.
Concluding remarks
This study demonstrates that the rational design of a multi-step functional screening procedure to guide sequencing is a very powerful strategy to accelerate enzyme discovery in metagenomes. Here, it was applied to identify highly prevalent genes encoding enzymes that are involved in the catabolism of the dietary fibers by the human gut microbiome and provided new insights into the gastrointestinal tract functional trophic chain. Besides, our procedure appears to efficiently identify clusters of potentially complementary activities for the complete breakdown of complex plant polysaccharides, which can be of prime interest for biorefinery processes and white biotechnologies. Their potential for such applications will have to be evaluated in future works. Finally, we note that the strategy reported here, which coupled functional screens and sequence-based metagenomics, is highly generic and can be applied to mine other ecosystems known to be highly specialized for raw biomass degradation (i.e., rumen and insect gut microbiomes) for novel biocatalysts.
Methods
Construction of the metagenomic library
The fecal sample was collected from a healthy 30-yr-old male who followed a vegetarian and fish-eating diet. His ascendants were omnivorous. The individual did not eat any functional food such as prebiotics or probiotics, nor did he receive any antibiotics or other drugs during the 6 mo before sampling. The bacterial fraction was recovered from 2 g of feces by a gradient density technique using Nycodenz as previously described (Courtois et al. 2003). The bacterial cell fraction was collected, washed with ultra-pure water, then centrifuged for 10 min at 12,000g. The cell pellet was resuspended in a 50 mM Tris (pH 8), 100 mM EDTA buffer and then incorporated in low-melt-point agarose before a gentle enzymatic lysis, as described by Ginolhac et al. (2004). High-molecular-weight bacterial DNA trapped in agarose plugs was immediately inserted into the wells of a 0.8% low-melting-temperature gel (Bio-Rad) and separated for 18 h by pulsed-field gel electrophoresis at 4.5 V/cm with 5- to 40-sec pulse times with a CHEFDRIII apparatus (Bio-Rad). DNA fragments with size ranging from 30 to 40 kb were isolated and recovered from the gel with GELase (Epicentre Technologies). Phylogenetic analysis of the extracted metagenomic DNA using 16S rDNA sequencing was performed according to Tap et al. (2009). The GenBank accession numbers for the 16S rDNA molecular inventory are HM475513–HM480042. The correspondence between the bacterial clone numbers appearing in Supplemental Figure S2 and the corresponding GenBank accession numbers is mentioned in Supplemental Table S5.
The metagenomic DNA was then cloned into fosmids by using the pCC1FOS fosmid library production kit (Epicentre Technologies) as recommended by the manufacturer. Recombinant colonies were transferred to 384-well microtiter plates containing freezing medium (Luria-Bertani, 8% glycerol complemented with 12.5 g/mL chloramphenicol), using an automated colony picker (QpixII; Genetix). After 22 h of growth at 37°C without any agitation, the plates were stored at −80°C.
High-throughput functional screens
Metagenomic clones were screened for polysaccharide digestion activities by spotting them on 22 cm × 22 cm bioassay trays containing solid agar and the target polysaccharide, using a QPixII (Genetix) colony picker. Solid agar was either PLA (agar-supplemented LB buffered to pH 6.6 by addition of 5.4 g/L Na2HPO4·12H2O and 4.8 g/L NaH2PO4·H2O) or, in the case of starch related polysaccharide containing media, terrific broth (TB). All media were supplemented with 12.5 mg/L chloramphenicol and with polysaccharides (beta-glucans, xylans, pectin, amylose, galactan) as listed in Supplemental Table S1. The assay plates were incubated for 7 d at 37°C, except for plates containing AZCL-amylose, which were incubated for only 3 d to avoid interference with E. coli host starch-degrading activities. A final throughput of 200,000 clones assayed per week and per substrate was achieved.
After incubation on plates containing chromogenic polysaccharides, positive clones were visually detected by the presence of a blue or red halo resulting from the production of colored oligosaccharides that diffused around the bacterial colonies. For pectin assays, the plates were colored for 20 min with an aqueous solution of Ruthenium Red (0.5% m/v) at room temperature. After removing exceeding Ruthenium Red solution by aspiration, clear halos were observed around the positive clones.
Secondary screens
All positive clones were further screened for hydrolysis efficiency and specificity toward various polysaccharide structures, by screening them on solid agar containing polysaccharides of various structures (Supplemental Table S1). Native polysaccharides were added to the sterile agar media at 50°C to conserve their crystalline structure. Ten microliters of overnight liquid cultures of the positive clones were placed on the agar surface, and the plates were incubated for 3 to 7 d at 37°C. Plates containing nonchromogenic beta-glucans and xylan were stained with an aqueous solution of Congo Red (0.05% m/v) followed by an overnight exposure to 1 M NaCl. Digestion zones were visible as clear halos around the positive colonies, except the deep brown halos observed for carboxymethyl cellulose. Nonchromogenic amylose- (Potocki-Veronese et al. 2005) and starch-containing plates were stained by exposure to iodine vapor, revealing unstained halos around positive colonies. Nonchromogenic pectic polysaccharides were stained with Ruthenium Red as described in the previous section.
To measure enzyme thermostability and activities at various pH values, positive clones were grown in liquid cultures in 96-well microplates. Cell lysis was performed by addition of 0.5 mg/mL lysozyme and one cycle of freeze/thaw at −20°C. For thermostability assays, cell extracts were incubated for 15 min at 55°C. Cell extracts were incubated in 20 mM citrate-phosphate buffer at pH 4, 7, and 9, supplemented with 0.1% AZCL-polysaccharides (same as used in primary screens), for 24 h at 37°C. Polysaccharide hydrolysis resulting in the release of soluble blue oligosaccharides was quantified by measuring absorbance at 590 nm.
Transposon mutagenesis of the DNA inserts from the 26 selected clones was performed using the EZ-Tn5 <oriV/KAN-2> Insertion Kit (Epicentre). Inactivated clones were identified by plating isolated colonies on agar-supplemented LB containing 12.5 mg/L chloramphenicol, 50 mg/L kanamycine, and the polysaccharide used in the primary screens. Sanger sequencing was performed outward from the nested transposon using the primers supplied in the kit.
Pyrosequencing, read assembly, and gene prediction
Pyrosequencing of whole fosmid inserts was performed on a 454 Life Sciences (Roche) GS FLX system by the Genoscope sequencing facility (Evry, France), yielding in total 186,762 contigable reads. Read assembly was done using CAP3 (Huang and Madan 1999), a DNA Sequence Assembly Program, and resulted in 106 contigs sizing between 113 bp and 51,798 bp, covering in total 1,002,117 bp. Ninety-eight percent of the sequenced nucleotides were included in 27 large contigs of at least 8343 nt, obtained with a mean sequencing depth of 44×. Two large contigs were found for clone 4. These 27 large contigs were further used for analysis. pCC1FOS sequences were identified using Crossmatch (http://bozeman.mbt.washington.edu/phredphrapconsed.html), discarded, and replaced by NNN. Excluding the vector sequences, these 27 large contigs included 881,473 nt of metagenomic DNA. The comparison of these sequences with themselves revealed three cases of partial sequence redundancy, which always occurred between clones presenting the same enzymatic activity detected using the primary screens. In the first two cases, the 5′ extremity of a contig was identical to the 3′ extremity of a contig from another clone (clones 14/15 and 17/18), which allowed manual assembly of them to provide up to 71.3 kb of metagenomic DNA issued from one unique gut bacterium. In the case of beta-glucanase active clones, one sequence fragment (20.9 kb) from clone 10 was also found in the contig sequence from clone 11, without any homologies of the contig extremities. As described in this report, this particular sequence redundancy phenomenon may be due to HGTs. The Metagene program (http://metagene.cb.k.u-tokyo.ac.jp/metagene) was used to predict open reading frames (ORFs ≥ 20 amino acids) from the resulting sequences. No frameshift was detected in the gene sequences by using BLASTX comparison to the Uniref100 database, reflecting the reliability of read assembly and gene detection. For each of the 26 clones, the large contig sequence has been deposited in DDBJ/EMBL/GenBank under accession numbers GU942928–GU942942 and GU942944–GU942954.
ORF analysis
COG assignment of predicted gene products was made using RPS-BLAST analysis against the reference GOG data set. COG assignment was taken into account only for E-values ≤ 10−8. When a predicted gene product was assigned to multiple COGs, this hit was counted as divided by the number of assigned COGs, and the value was dispensed evenly to each COG. Signal peptide prediction was performed using PHOBIUS (http://www.ebi.ac.uk/Tools/phobius/). CAZyme-encoding genes were identified by BLAST analysis of the nucleotide sequences from the 106 contigs against the amino acid sequences derived from the CAZy database (http://www.cazy.org) using a cut-off E-value of 7 × 10−6. Other genes were manually annotated using NCBI-BLASTP against the NR database (E-value < 10−8, identity > 35%, query length coverage ≥ 50%). Gene prevalence in the human gut microbiome was detected by using a TBLASTN comparison of the protein sequences identified in this study to the metagenomic data sets available for 124 European (Qin et al. 2010), 13 Japanese (Kurokawa et al. 2007), and 46 U.S. individuals (Gill et al. 2006; Turnbaugh et al. 2009, 2010) (E-value = 0, identity ≥ 90% or identity = 100%).
Taxonomic assignment of metagenomic sequences
Two methodologies were used. The first was based on protein sequence similarities with proteins of sequenced genomes, using a BLASTP analysis against the nonredundant protein sequence database of the NCBI. For each protein of each metagenomic DNA fragment, the microbial origin of the best BLAST hit was assigned only for matches covering at least 50% of the protein length, with an E-value better than 10−8 and an identity of at least 90%. Proteins that did not pass those criteria were assigned to the “no hits” category. We assigned a class, genus, or species to the DNA fragment issued from one clone when at least 50% of the putative proteins encoded by this fragment presented a best BLAST hit issued from the same microbe. Also, if putative proteins encoded from the same DNA fragment had the best BLAST hit issued from microbes of different classes, we considered the entire fragment as unassignable. The second approach was based on tetranucleotide frequency count, an analysis related to genomic signatures, by using Ocount software (Teeling et al. 2004) connected to a previously designed pipeline allowing a normalization of tetranucleotide frequency according to sequence length (Tap et al. 2009). The 26-fosmid insert sequences were analyzed as divided into 10-kb fragments. Genetic diversity, recorded as 256-tetranucleotide distribution, was represented by a principle component analysis (PCA) using R software (Chessel et al. 2004). Only the first two PCA components, representing 49.7% of the total genetic diversity, were used to illustrate this analysis.
Acknowledgments
The high-throughput screening work was performed at the Laboratory for BioSystems & Process Engineering (Toulouse, France) with the ICEO automated facility. ICEO is supported by grants from the Région Midi-Pyrénées, France, the European Regional Development Fund, and the Institut National de la Recherche Agronomique, France (the French National Institute for Agricultural Research). We thank Sophie Bozonnet and Sandrine Laguerre for their assistance. This work was carried out with the financial support of the ANR—Agence Nationale de la Recherche—The French National Research Agency under the Programme National de Recherche en Alimentation et nutrition humaine, project ANR-06-PNRA-024. Pyrosequencing was funded by the French National Institute for Agricultural Research.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) under accession nos. GU942928–GU942942 and GU942944–GU942954.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.108332.110.
References
- Brennan Y, Callen WN, Christoffersen L, Dupree P, Goubet F, Healey S, Hernandez M, Keller M, Li K, Palackal N, et al. 2004. Unusual microbial xylanases from insect guts. Appl Environ Microbiol 70: 3609–3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Bagdasarian M, Kaufman MG, Bates AK, Walker ED 2007. Mutational analysis of the ompA promoter from Flavobacterium johnsoniae. J Bacteriol 189: 5108–5118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chessel D, Dufour AB, Thioulouse J 2004. The ade4 package—I: One-table methods. R News 4: 5–10 [Google Scholar]
- Courtois S, Cappellano CM, Ball M, Francou FX, Normand P, Helynck G, Martinez A, Kolvek SJ, Hopke J, Osburne MS, et al. 2003. Recombinant environmental libraries provide access to microbial diversity for drug discovery from natural products. Appl Environ Microbiol 69: 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan CJ, Xian L, Zhao GC, Feng Y, Pang H, Bai XL, Tang JL, Ma QS, Feng JX 2009. Isolation and partial characterization of novel genes encoding acidic cellulases from metagenomes of buffalo rumens. J Appl Microbiol 107: 245–256 [DOI] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA 2005. Diversity of the human intestinal microbial flora. Science 308: 1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Duan CJ, Pang H, Mo XC, Wu CF, Yu Y, Hu YL, Wei J, Tang JL, Feng JX 2007. Cloning and identification of novel cellulase genes from uncultured microorganisms in rabbit cecum and characterization of the expressed cellulases. Appl Microbiol Biotechnol 75: 319–328 [DOI] [PubMed] [Google Scholar]
- Ferrer M, Golyshina OV, Chernikova TN, Khachane AN, Reyes-Duarte D, Santos VA, Strompl C, Elborough K, Jarvis G, Neef A, et al. 2005. Novel hydrolase diversity retrieved from a metagenome library of bovine rumen microflora. Environ Microbiol 7: 1996–2010 [DOI] [PubMed] [Google Scholar]
- Ferrer M, Beloqui A, Timmis KN, Golyshin PN 2009. Metagenomics for mining new genetic resources of microbial communities. J Mol Microbiol Biotechnol 16: 109–123 [DOI] [PubMed] [Google Scholar]
- Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA 2008. Polysaccharide utilization by gut bacteria: Potential for new insights from genomic analysis. Nat Rev Microbiol 6: 121–131 [DOI] [PubMed] [Google Scholar]
- Gabor EM, Alkema WB, Janssen DB 2004. Quantifying the accessibility of the metagenome by random expression cloning techniques. Environ Microbiol 6: 879–886 [DOI] [PubMed] [Google Scholar]
- Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE 2006. Metagenomic analysis of the human distal gut microbiome. Science 312: 1355–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginolhac A, Jarrin C, Gillet B, Robe P, Pujic P, Tuphile K, Bertrand H, Vogel TM, Perriere G, Simonet P, et al. 2004. Phylogenetic analysis of polyketide synthase I domains from soil metagenomic libraries allows selection of promising clones. Appl Environ Microbiol 70: 5522–5527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabitske HA, Slavin JL 2008. Low-digestible carbohydrates in practice. J Am Diet Assoc 108: 1677–1681 [DOI] [PubMed] [Google Scholar]
- Grant S, Sorokin DY, Grant WD, Jones BE, Heaphy S 2004. A phylogenetic analysis of Wadi el Natrun soda lake cellulase enrichment cultures and identification of cellulase genes from these cultures. Extremophiles 8: 421–429 [DOI] [PubMed] [Google Scholar]
- Guo H, Feng Y, Mo X, Duan C, Tang J, Feng J 2008. [Cloning and expression of a beta-glucosidase gene umcel3G from metagenome of buffalo rumen and characterization of the translated product]. Sheng Wu Gong Cheng Xue Bao 24: 232–238 [PubMed] [Google Scholar]
- Huang X, Madan A 1999. CAP3: A DNA sequence assembly program. Genome Res 9: 868–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Institute of Medicine 2005. Dietary reference intakes. National Academy of Sciences, Washington, DC [Google Scholar]
- Jones BV, Marchesi JR 2007. Transposon-aided capture (TRACA) of plasmids resident in the human gut mobile metagenome. Nat Methods 4: 55–61 [DOI] [PubMed] [Google Scholar]
- Kalyuzhnaya MG, Lapidus A, Ivanova N, Copeland AC, McHardy AC, Szeto E, Salamov A, Grigoriev IV, Suciu D, Levine SR, et al. 2008. High-resolution metagenomics targets specific functional types in complex microbial communities. Nat Biotechnol 26: 1029–1034 [DOI] [PubMed] [Google Scholar]
- Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, Takami H, Morita H, Sharma VK, Srivastava TP, et al. 2007. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res 14: 169–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, et al. 2008. Evolution of mammals and their gut microbes. Science 320: 1647–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LL, McCorkle SR, Monchy S, Taghavi S, van der Lelie D 2009. Bioprospecting metagenomes: Glycosyl hydrolases for converting biomass. Biotechnol Biofuels 2: 10 doi: 10.1186/1754-6834-2-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JR, Duan CH, Zhao X, Tzen JT, Cheng KJ, Pai CK 2008. Cloning of a rumen fungal xylanase gene and purification of the recombinant enzyme via artificial oil bodies. Appl Microbiol Biotechnol 79: 225–233 [DOI] [PubMed] [Google Scholar]
- Lozupone CA, Hamady M, Cantarel BL, Coutinho PM, Henrissat B, Gordon JI, Knight R 2008. The convergence of carbohydrate active gene repertoires in human gut microbes. Proc Natl Acad Sci 105: 15076–15081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald TT, Monteleone G 2005. Immunity, inflammation, and allergy in the gut. Science 307: 1920–1925 [DOI] [PubMed] [Google Scholar]
- Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK, et al. 2009. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci 106: 5859–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, et al. 2006. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 55: 205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Koropatkin NM, Smith TJ, Gordon JI 2009. Complex glycan catabolism by the human gut microbiota: The Bacteroidetes Sus-like paradigm. J Biol Chem 284: 24673–24677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarr SE, Ridlon JM, Hylemon PB 2005. Diet, anaerobic bacterial metabolism, and colon cancer: A review of the literature. J Clin Gastroenterol 39: 98–109 [PubMed] [Google Scholar]
- Pang H, Zhang P, Duan CJ, Mo XC, Tang JL, Feng JX 2009. Identification of cellulase genes from the metagenomes of compost soils and functional characterization of one novel endoglucanase. Curr Microbiol 58: 404–408 [DOI] [PubMed] [Google Scholar]
- Potocki-Veronese G, Putaux JL, Dupeyre D, Albenne C, Remaud-Simeon M, Monsan P, Buleon A 2005. Amylose synthesized in vitro by amylosucrase: Morphology, structure, and properties. Biomacromolecules 6: 1000–1011 [DOI] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464: 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu A, Brulc JM, Wilson MK, Law BF, Theoret JR, Joens LA, Konkel ME, Angly F, Dinsdale EA, Edwards RA, et al. 2008. Comparative metagenomics reveals host specific metavirulomes and horizontal gene transfer elements in the chicken cecum microbiome. PLoS ONE 3: e2945 doi: 10.1371/journal.pone.0002945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R 2004. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118: 229–241 [DOI] [PubMed] [Google Scholar]
- Rees HC, Grant S, Jones B, Grant WD, Heaphy S 2003. Detecting cellulase and esterase enzyme activities encoded by novel genes present in environmental DNA libraries. Extremophiles 7: 415–421 [DOI] [PubMed] [Google Scholar]
- Richardson TH, Tan X, Frey G, Callen W, Cabell M, Lam D, Macomber J, Short JM, Robertson DE, Miller C 2002. A novel, high performance enzyme for starch liquefaction. Discovery and optimization of a low pH, thermostable alpha-amylase. J Biol Chem 277: 26501–26507 [DOI] [PubMed] [Google Scholar]
- Roberts AP, Chandler M, Courvalin P, Guedon G, Mullany P, Pembroke T, Rood JI, Smith CJ, Summers AO, Tsuda M, et al. 2008. Revised nomenclature for transposable genetic elements. Plasmid 60: 167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondon MR, August PR, Bettermann AD, Brady SF, Grossman TH, Liles MR, Loiacono KA, Lynch BA, MacNeil IA, Minor C, et al. 2000. Cloning the soil metagenome: A strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl Environ Microbiol 66: 2541–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvendran RR 1984. The plant cell wall as a source of dietary fiber: Chemistry and structure. Am J Clin Nutr 39: 320–337 [DOI] [PubMed] [Google Scholar]
- Simon C, Daniel R 2009. Achievements and new knowledge unraveled by metagenomic approaches. Appl Microbiol Biotechnol 85: 265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI 2005. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307: 1955–1959 [DOI] [PubMed] [Google Scholar]
- Tamames J, Moya A 2008. Estimating the extent of horizontal gene transfer in metagenomic sequences. BMC Genomics 9: 136 doi: 10.1186/1471-2164-9-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang K, Utairungsee T, Kanokratana P, Sriprang R, Champreda V, Eurwilaichitr L, Tanapongpipat S 2006. Characterization of a novel cyclomaltodextrinase expressed from environmental DNA isolated from Bor Khleung hot spring in Thailand. FEMS Microbiol Lett 260: 91–99 [DOI] [PubMed] [Google Scholar]
- Tang K, Kobayashi RS, Champreda V, Eurwilaichitr L, Tanapongpipat S 2008. Isolation and characterization of a novel thermostable neopullulanase-like enzyme from a hot spring in Thailand. Biosci Biotechnol Biochem 72: 1448–1456 [DOI] [PubMed] [Google Scholar]
- Tap J, Mondot S, Levenez F, Pelletier E, Caron C, Furet JP, Ugarte E, Munoz-Tamayo R, Paslier DL, Nalin R, et al. 2009. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol 11: 2574–2584 [DOI] [PubMed] [Google Scholar]
- Teeling H, Waldmann J, Lombardot T, Bauer M, Glockner FO 2004. TETRA: A web-service and a stand-alone program for the analysis and comparison of tetranucleotide usage patterns in DNA sequences. BMC Bioinformatics 5: 163 doi: 10.1186/1471-2105-5-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temperton B, Field D, Oliver A, Tiwari B, Muhling M, Joint I, Gilbert JA 2009. Bias in assessments of marine microbial biodiversity in fosmid libraries as evaluated by pyrosequencing. ISME J 3: 792–796 [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Gordon JI 2009. The core gut microbiome, energy balance and obesity. J Physiol 587: 4153–4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. 2009. A core gut microbiome in obese and lean twins. Nature 457: 480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Quince C, Faith JJ, McHardy AC, Yatsunenko T, Niazi F, Affourtit J, Egholm M, Henrissat B, Knight R, et al. 2010. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Natl Acad Sci 107: 7503–7508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama T, Miyazaki K 2009. Functional metagenomics for enzyme discovery: Challenges to efficient screening. Curr Opin Biotechnol 20: 616–622 [DOI] [PubMed] [Google Scholar]
- Voget S, Leggewie C, Uesbeck A, Raasch C, Jaeger KE, Streit WR 2003. Prospecting for novel biocatalysts in a soil metagenome. Appl Environ Microbiol 69: 6235–6242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization 2003. Diet, nutrition and the prevention of chronic disease. Technical Report Series no. 916. http://whqlibdoc.who.int/trs/who_TRS_916.pdf [PubMed]
- Zoetendal EG, Akkermans AD, De Vos WM 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol 64: 3854–3859 [DOI] [PMC free article] [PubMed] [Google Scholar]