Abstract
Proteorhodopsin (PR), a photoactive proton pump containing retinal, is present in approximately half of all bacteria in the ocean, but its physiological role is still unclear, since very few strains carrying the PR gene have been cultured. The aim of this work was to characterize PR diversity in a North Sea water sample, cultivate a strain representative of North Sea PR clusters, and study the effects of light and carbon concentration on the expression of the PR gene. A total of 117 PR sequences, of which 101 were unique, were obtained from a clone library of PCR-amplified PR gene fragments. Of the North Sea PRs, 97% were green light absorbing, as inferred from the amino acid at position 105; 67% of the PR protein fragments showed closest similarity to PRs from Alphaproteobacteria, 4% showed closest similarity to PRs from Gammaproteobacteria, and 29% showed closest similarity to PRs from “Bacteroidetes”/Flavobacteria. The dominant PR cluster (comprising 18% of all PRs) showed a high degree of similarity to the PR from the cultivated Roseobacter strain HTCC2255. The relative abundances of the North Sea PR clusters were confirmed by quantitative PCR. They were detected in metagenomic fragments from coastal oceans worldwide with various degrees of abundance. Several hundred bacterial strains from the North Sea water sample were cultivated on oligocarbophilic media. By screening with degenerate primers, two strains carrying the PR gene were identified. Their 16S rRNA gene sequences were identical and affiliated with a Bacteroidetes subcluster from the North Sea. The PR sequence of isolate PRO95 was completed by chromosomal walking. It was 76% identical to that of Dokdonia donghaensis MED134 and was functional, as indicated by the signature amino acids. PRO95 expressed its PR gene in liquid media containing between 9.7 and 121 mM carbon, both in the light and in the dark. Growth was not enhanced by light. Thus, the detection of the physiological role of PR may require more sensitive methods.
Proteorhodopsins (PRs) are retinal-binding integral membrane proteins belonging to the microbial rhodopsin superfamily (36) and are suggested to provide energy for microbial metabolism in marine ecosystems by using a light-generated proton gradient for photophosphorylation (17, 18, 26). They were first discovered by analysis of the genome fragment of a marine gammaproteobacterium from the SAR86 clade (3, 4). Since then, PRs belonging to different bacterial groups, e.g., the ubiquitous alphaproteobacterial SAR11 clade and Roseobacter and “Bacteroidetes” species, have been found in many different marine environments (6, 9, 25, 27, 29-33, 41, 41, 45), as well as in freshwater and brackish ecosystems (2). PR has also been found in marine planktonic Archaea (11). It is currently estimated that up to 70% of the bacteria in the Sargasso Sea, between 7 and 57% of bacteria elsewhere in the North Atlantic Ocean, and about 13% of the bacteria in the Mediterranean Sea contain PR (6, 29, 32). It has been shown previously that a single amino acid substitution in the PR protein sequence leads to a drastic change in the light absorption maximum from 490 nm (blue) to 520 to 540 nm (green) (25). The natural occurrence and vertical distribution of green- versus blue-light-absorbing PRs in the oceans can therefore at least partly be accounted for by adaptation to the prevailing light spectra (29, 31). PRs from uncultivated marine bacteria have been expressed in Escherichia coli and shown to transport protons into the surrounding medium upon illumination (3) by a mechanism similar to that of bacteriorhodopsin in Archaea (28). The resulting proton gradient can be used by E. coli to generate ATP (26) or to power the flagellar motor (42).
All these data strongly suggest that PRs provide an adaptive advantage to the bacteria in the ocean. However, very few PR-containing bacteria have been successfully cultured, and for those that have been investigated, no clear conclusion regarding the metabolic role of PR has yet been drawn (12). With one exception, no growth advantage for the bacteria in the light compared to the dark could be demonstrated. Neither “Candidatus Pelagibacter ubique,” an alphaproteobacterium from the abundant marine SAR11 clade (14), nor the gammaproteobacterium HTCC2207 from the SAR92 clade (37) shows higher growth yields or higher growth rates when incubated in the light. The role of xanthorhodopsin in strain HTCC2181, a member of the OM43 clade of marine betaproteobacteria, is currently unknown (15). The flavobacterium Polaribacter dokdonensis MED152 (18) also does not grow better in the light; however, the amount of bicarbonate fixed through anaplerotic enzymes is larger in the light than in the dark. This process may allow MED152 to more efficiently use organic matter for biosynthesis in the light. Finally, Dokdonia donghaensis MED134 (17) is the only strain studied so far which shows higher growth yields in the light, but only at very low carbon concentrations, between 0.1 and 1.1 mM. No difference in growth in full-strength medium (242 mM carbon) is seen (17).
Thus, there is a need for cultivating marine bacteria which produce PR and are representative of abundant marine clades. By studying them under ecologically relevant conditions, the physiological role (or roles) of their light-dependent proton pump PR may be unraveled. The aim of this study was to isolate a PR-producing strain from a North Sea water sample which is representative of the prevailing natural microbial community and to examine the effects of carbon concentration and light on the expression of the PR gene of this strain.
MATERIALS AND METHODS
Sampling and extraction of total DNA.
A water sample (5 liters) from the Kabeltonne site at Helgoland Roads in the North Sea (54°11.18′N, 7°54′E) was collected at a depth of 0.5 m in September 2006 and processed immediately at the Biological Station of Helgoland. The water sample was filtered first through a 10-μm-pore-size filter and then through a 0.2-μm-pore-size filter. Total DNA was extracted from the 0.2-μm filter with the FastDNA Spin kit for soil (Qbiogene) and used for the clone library. An aliquot (10 ml) of the filtrate from the 10-μm filter was removed and immediately used for isolation of bacteria (see below).
PCR amplification, cloning, and sequencing.
Primers used in this study are listed in Table 1.
TABLE 1.
Primers used in this study
| Name | Sequence (5′-3′) | Taa (°C) | Product size | Applicationc | Reference |
|---|---|---|---|---|---|
| Primers for PR genes | |||||
| PR1 | MGN TAY ATH GAY TGG YT | 47 | 312 bp | Scr | 32 |
| PR2 | WWN MGN TAY GTN GAY TGG | ||||
| PR3 | GGR TAD ATN GCC CAN CC | ||||
| PR4 | GTC AGG CTG GTC TTA CGT AGA CTG | 66 | NAb | Chrw | This study |
| PR5 | TTA CTC GCC GAC CCT ACA AAG CTA | 65 | NA | Chrw | This study |
| PRO95_REV_68 | GCA GAA GCT GCC ATC ATA GC | 62 | NA | Chrw | This study |
| PRO95_REV_395 | ATA GCT CCC CAT AAT GCT GC | 60 | NA | Chrw | This study |
| PRO95_PR_F | CCC CAT ATG AAA TTT TTA TTA C | 53 | 756 bp | Con | This study |
| PRO95_PR_R | CCC CAT ATG TTA AAC AAC C | 56 | |||
| PRO95_FOR | CGT TCC ATT AAT GTG CGT TG | 59 | 158 bp | qRT-PCR | This study |
| PRO95_REV | TAG CTC CCC ATA ATG CTG CT | ||||
| SARPR_125F | THG GWG GAT AYT TAG GWG AAG C | 54 | 163 bp | qPCR | 6 |
| SARPR_288R | CCC AAC CWA YWG TWA CRA TCA TTC T | ||||
| Flavo_F2 | GGA TAC TAA CAG TTC CWT TRA TGT GTG | 56 | qPCR | 6 | |
| Flavo_R2 | CCR AAG TAA CCW GTR ACA AGC AT | ||||
| HTCC2255_F | ATG CTC GTA GGT GGG TAT GC | 55 | qPCR | This study | |
| HTCC2255_R | GCC TCA TCT GCA GCA ACT TT | ||||
| MEDHO_f | TGC CGC AGT CTC YTC WAG TTC | 55 | qPCR | This study | |
| MEDHO_r | GAA TCC TAG GCT WGC YGA CAT | ||||
| MS0242_F | TTA CTG TGC CAT TAA TGT GTG TTG | 55 | qPCR | This study | |
| MS0242_R | AAT CCA GAA GTA TGC AAG ACC TG | ||||
| Flavo-H_f | CTA ACW GTD CCW TTR ATG TGT GTT G | 55 | qPCR | This study | |
| Flavo-H_r | CCT GTW GTG TAW ACW GCT TCT CC | ||||
| Primers for 16S rRNA genes | |||||
| 16F27 | AGA GTT TGA TCC TGG CTC AG | 54 | 1.5 kb | Phy | 22 |
| 16R1492 | TAC GGY TAC CTT GTT ACG ACT T | ||||
| BACT1369F | CGG TGA ATA CGT TCY CGG | 58 | 174 bp | qPCR | 38 |
| PROK1543R | AAG GAG GTG ATC CR GCC GCA | ||||
| PRO95_16SF | TGC GAT AGA TAG GGG TCC TG | 59 | 150 bp | qRT-PCR | This study |
| PRO95_16SR | ACC CAT AGG GCA GTC TTC CT |
Ta, annealing temperature.
NA, not applicable.
Application of the primer: scr, screening; chrw, chromosomal walking; con, confirmation of PR gene sequence (amplification of the complete PR gene from D. donghaensis PRO95); qPCR, quantitative PCR; and phy, phylogenetic analysis.
(i) PR gene fragment.
For detection of PR genes in environmental DNA or genomic DNA samples from isolates, a multiplex PCR analysis with degenerated primers (PR1, PR2, and PR3 [Table 1]) was performed. PCRs for PR were performed using Taq DNA polymerase (Qiagen). PCR amplification was carried out with a total volume of 20 μl containing ∼10 ng of template DNA, 200 μM (each) deoxynucleoside triphosphates (dNTPs), 1 μM (each) primers, and 0.5 U of Taq DNA polymerase. The amplification comprised the following program: an initial step at 94°C for 1 min and then 35 cycles at 94°C for 10 s, 47°C for 30 s, and 68°C for 50 s. At the end of every PCR, a postelongation step at 68°C for 90 s was carried out. PCR products were visualized by gel electrophoresis. The product (312 bp) was excised from the gel and purified with the QIAquick gel extraction kit (Qiagen). The purified DNA fragments were cloned with the TOPO TA cloning kit for sequencing (Invitrogen) and sequenced. Nucleotide sequences were assembled in the forward direction and read in reverse.
(ii) Complete PR gene.
For confirmation of the complete assembled PR gene sequence, primers PRO95_PR_F and PRO95_PR_R (Table 1) were used. PCR amplification was carried out as described above. The product (756 bp, including the 741-bp PR gene sequence and an NdeI restriction site on each end) was cloned into the pTrcHis-TOPO vector (Invitrogen) and sequenced.
(iii) 16S rRNA gene.
The PCR for the 16S rRNA gene was also performed in the same way, except that 0.5 μM primers and 2 U of Taq DNA polymerase were used; the amplification comprised the following program: an initial step at 94°C for 2 min and then 30 cycles at 94°C for 1 min, 54°C for 30 s, and 72°C for 2 min. Postelongation was carried out at 72°C for 10 min. All further steps were done in the same way as those for the partial PR gene.
Estimation of sampling efficiency.
Nucleotide sequences were subjected to multiple-sequence alignment analysis using the ClustalX program (40). A DNA distance matrix was generated using DNADIST from the PHYLIP package (http://evolution.genetics.washington.edu/phylip.html) with Jukes-Cantor correction for multiple substitutions per site. Based on this matrix, the program DOTUR (35) was used for clustering sequences into operational taxonomical units (OTUs) by using the farthest-neighbor algorithm. For unique sequences and sequences with differences of 6 and 15%, rarefaction curves estimating the sampling progress and collector's curves estimating the OTU richness (7) at the sampling site were constructed. For clustering of protein sequences with DOTUR, a distance matrix generated with PROTDIST from the PHYLIP package was used.
Phylogenetic analysis of PR sequences.
The PR amino acid sequence was identified using ORF Finder (http://www.bioinformatics.org/sms/index.html) and BLAST (1). Phylogenetic relationships were inferred from about 90 amino acid positions (excluding the regions corresponding to primers) by the neighbor-joining method (34). The evolutionary distances were computed by the Poisson correction method (46) and are expressed as numbers of amino acid substitutions per site. All positions corresponding to alignment gaps and missing data were eliminated only in pairwise sequence comparisons. Bootstrap sampling (with 1,000 replicates) was used for validation. Phylogenetic analyses were conducted with MEGA4 software (39). For visualization of the obtained phylogenetic tree, the Interactive Tree of Life (iTOL) tool was used (23).
Estimation of PR abundance by quantitative PCR.
Primers used for quantitative PCR analysis are listed in Table 1. Amplification efficiencies of the primer pairs, determined with serial dilutions of sample DNA, were as follows: for 16S rRNA genes, 1.82; for SAR11 PR sequences, 1.67; for HTCC2255 PR sequences, 1.92; for MEDHO1202F4 PR sequences, 1.81; for MS0242A PR sequences, 1.78; for flavobacterial subset NASB (Flavo-NASB) PR sequences, 1.56; and for Helgoland flavobacterial (Flavo-HEL) PR sequences, 1.52. For relative quantification, 1 ng of DNA was used as a template. Quantitative PCR was performed in quadruplicate with a LightCycler 480 system (Roche) and a QuantiTect Sybr green PCR kit (Qiagen): initial denaturation at 95°C for 15 min was followed by 60 cycles at 94°C for 10 s, the annealing temperature indicated in Table 1 for 30 s, and 72°C for 30 s. Melting curve analysis followed the amplification steps. The ratio of 1.9 rRNA genes to the average of 6 single-copy genes was determined in the Sargasso Sea metagenomic study (41). We used this ratio to normalize PR gene abundance with respect to the abundance of 16S rRNA genes.
Biogeographic analysis.
A BLAST search comparing all Helgoland PR sequences against the Global Ocean Sampling (GOS) expedition, Monterey Bay, Botany Bay, and Antarctica metagenomic libraries obtained from the Community Cyberinfrastructure for Advanced Marine Microbial Ecology Research and Analysis (CAMERA; http://camera.calit2.net/) was performed. A description of the sampling sites can be found at the CAMERA website. An E value of 10−30, corresponding to a sequence identity of roughly 80%, was used as a cutoff (6). The obtained sequences were clustered due to their relationship to PRs from Bacteroidetes, Gammaproteobacteria, and the alphaproteobacterial SAR11, HTCC2255, and MedPR13e07 clusters.
Isolation and cultivation of bacterial strains and DNA extraction.
The water sample analyzed for PR diversity (see above) had first been filtered through a 10-μm filter. An aliquot of this filtrate (10 ml, containing particles of <10 μm) was removed and used immediately for cultivation of bacteria. It was serially diluted in sterile North Sea water that had been passed through a 0.2-μm filter. Oligocarbophilic bacteria were isolated by plating 50-μl aliquots of 1:10, 1:20, 1:40, and 1:100 dilutions in triplicate onto solid medium containing only autoclaved, sterile filtered water from the North Sea and agar (1.5%). A total of 260 plates were inoculated and incubated at an in situ temperature (16°C) in a natural day-night cycle, in the light, or in the dark for 4 weeks. All single colonies (approximately 400) were picked, suspended in filtered, autoclaved North Sea water, and streaked onto master plates of solid marine agar 2216 (Difco) for purification. To make glycerol stocks, single colonies were picked and grown in liquid marine broth (MB) medium (Difco marine broth 2216) at room temperature. Chromosomal DNA from every isolate was extracted from 1 ml of MB culture using the NucleoSpin tissue kit (Macherey-Nagel).
Cultivation of PRO95.
Isolate PRO95 was routinely cultivated on plates of Difco marine agar 2216 at room temperature (20 to 25°C). Growth in liquid medium was studied using MB medium (Difco marine broth 2216), amended with sea salts (Sigma), at half strength (3 g nutrients liter−1, 121 mM carbon), 1/5 strength (1.2 g nutrients liter−1, 48.4 mM carbon), 1/10 strength (0.6 g nutrients liter−1, 24.2 mM carbon), and 1/25 strength (0.24 g nutrients liter−1, 9.7 mM carbon). Carbon sources in marine broth 2216 are peptone (5 g liter−1) and yeast extract (1 g liter−1). Sea salts were added to the diluted medium to obtain the total amount of salts (31.8 g liter−1) in full-strength marine broth 2216 medium. The carbon concentration was calculated as described in reference 17. (See File S2 in the supplemental material for additional details.) Cultures were shaken at 150 to 200 rpm in a natural day-night cycle, in the dark, and in constant light from a 75-W halogen bulb at a distance of 40 to 60 cm. Growth was evaluated by measuring the optical density (OD) at 600 nm.
Chromosomal walking.
To obtain the complete PR gene sequence of isolate PRO95, primers binding shortly before the end of the known sequence but directed toward the outside were designed and used for direct sequencing of the chromosomal DNA. Obtained fragments were assembled, and new primers farther outside the PR gene (PR4, PR5, PRO95_REV_68, and PRO95_REV_395 [Table 1]) were designed and used for a second round of chromosomal walking. Sequencing was performed with an ABI sequencer using the following reaction mixture: 10 μl DNA, 4 μl BigDye Terminator v3.1 reagent, 0.5 μl primer (10 pmol/μl), 2 μl sequencing buffer, and water to obtain a final volume of 20 μl. The PCR program consisted of an initial step at 96°C for 1 min, 99 cycles of 96°C for 10 s, 55°C for 5 min, and 60°C for 4 min, and a final cooling step at 4°C.
Alignment of PRO95 PR with other PR sequences.
The alignment of PR protein sequences was produced with ClustalW using the default parameters and the Gonnet protein weight matrix. Prediction of transmembrane regions was performed using TMpred (http://www.ch.embnet.org/software/TMPRED_form.html).
RNA extraction and purification.
Cells of PRO95 were cultured in different concentrations of MB medium (1/2, 1/5, 1/10, and 1/25 dilutions) amended with sea salts (see “Cultivation of PRO95” above). PRO95 liquid culture samples of 2 ml (for MB diluted 1/2), 4 ml (for MB diluted 1/5), 8 ml (for MB diluted 1/10), and 16 ml (for MB diluted 1/25) were mixed with RNAprotect bacterial reagent (Qiagen) in an amount double the sample volume, and the mixtures were centrifuged for 10 min at 8,000 × g and 15°C. Alternatively, cultures were centrifuged for 4 to 5 min at 8,000 × g and 4°C and then the pellets were immediately suspended in RNAprotect bacterial reagent. All quantitative reverse transcription-PCR (qRT-PCR) determinations were obtained by both methods in parallel, except for the measurement of PR expression in the light, which was determined using only the first method. After resuspension of the pellets, cells were centrifuged again for 10 min at 8,000 × g and 15°C. Pellets were lysed enzymatically with lysozyme (15 mg/ml in 1× Tris-EDTA buffer) for 45 min at room temperature. RNA was isolated using the RNeasy minikit (Qiagen). In addition to the on-column DNase treatment using the RNase-free DNase set (Qiagen), a second DNase digestion in solution, also done with the RNase-free DNase set (Qiagen), was performed as a final cleaning step. The RNA concentration was determined using a Nanodrop 1000 spectrometer.
cDNA synthesis.
The reaction mix contained 750 ng (2.5 μl) random primers, 1 μg total RNA, 10 mM dNTPs (1 μl), and sterile diethyl pyrocarbonate-treated water (added to obtain a final volume of 10.5 μl). The reaction mix was incubated at 70°C for 10 min and then at 25°C for 10 min. The contents of the reaction tube were collected by brief centrifugation, after which 4 μl 5× first-strand buffer, 2 μl 100 mM dithiothreitol, and 1 μl (40 U) RNaseOUT were added and the preparation was subjected to gentle mixing and incubation at room temperature (25°C) for 2 min. Five hundred units (2.5 μl) of SuperScript II reverse transcriptase (Invitrogen) was added to obtain a final volume of 20 μl. The reaction mix was incubated in the following steps: 10 min at 25°C, 1 h at 37°C, 1 h at 42°C, and 10 min at 70°C. Then 6.66 μl 1 N NaOH was added, and the mix was incubated at 65°C for 30 min and at 25°C for 10 min. Finally, 6.66 μl 1 N HCl was added to neutralize NaOH. cDNA was purified using the QIAquick PCR purification kit (Qiagen).
Analysis of PR expression by quantitative real-time RT-PCR.
Primers used for quantitative PCR analysis are listed in Table 1. The amplification efficiencies of the primer pairs were determined with serial dilutions of sample DNA. For relative quantification, 1 ng of cDNA was used as a template. Quantitative PCR was performed with a LightCycler 480 system (Roche) and a QuantiTect Sybr green PCR kit (Qiagen): initial denaturation at 95°C for 15 min was followed by 50 cycles at 94°C for 10 s, 55°C for 30 s, and 72°C for 30 s.
Nucleotide sequence accession numbers.
Sequences of cloned PR gene fragments were deposited in EMBL under the accession numbers FJ560751 to FJ560867. Sequences of the complete PR gene and the 16S rRNA gene of isolate PRO95 were deposited in EMBL under accession numbers FJ627053 and FJ627052, respectively.
RESULTS AND DISCUSSION
Characterization of PR diversity and spectral tuning in the bacterioplankton community of a North Sea water sample.
The sampling site Kabeltonne is located at Helgoland Roads, an anchorage area between the two islands at Helgoland, approximately 60 km off the coast in the North Sea. Here, one of the longest continuous series of data for any marine station is available. The first data were collected in 1872, and continuous measurements and samplings on a daily to weekly schedule have been performed since 1962, thus providing a unique resource for investigations of long-term trends (10). While extensive data on physical and chemical parameters, phytoplankton, zooplankton, and the diversity of microbial communities are available (13), nothing was known about PR.
PR gene diversity.
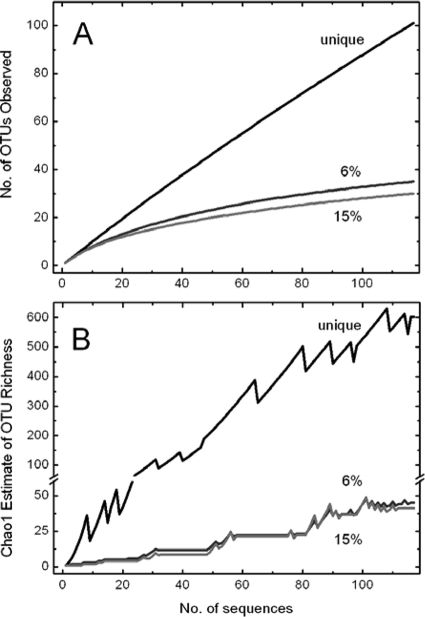
A wide diversity of PR variants was discovered in a near-surface water sample taken from the sampling site Kabeltonne. By PCR amplification using degenerated primers (32), cloning, and sequencing, 117 partial PR gene nucleotide sequences were obtained. Of these 117 sequences, 101 were unique. By using a species-specific cutoff of 6% sequence divergence, as calculated for pufM sequences from cultivated bacteria (44) and rpoD sequences from the Sargasso Sea metagenome (41), these sequences could be clustered into 35 OTUs by the DOTUR program. Divergence of 15%, calculated as a phylum-specific cutoff for pufM sequences (44), resulted in 30 OTUs. Rarefaction curves calculated for both cutoffs (Fig. 1A) did not reach saturation, indicating that the number of sequenced clones may not have been high enough to cover all species-specific PR sequences that could be amplified from the sample DNA with the primer combinations used. However, according to the Chao richness estimation (Fig. 1B), we could expect 38 to 71 and 33 to 70 OTUs (within a confidence interval of 95%) for 6 and 15% sequence divergences, respectively, meaning that our study covered at least half of the PR sequences at the tested diversity levels.
FIG. 1.
Rarefaction analysis (A) and Chao richness estimation (B) for PR sequences retrieved from the North Sea near the island of Helgoland. All 117 nucleotide sequences were grouped into OTUs based on 0% divergence (unique sequences) and 6 and 15% divergence cutoffs. Rarefaction curves and Chao richness estimation curves are labeled according to the cutoff value.
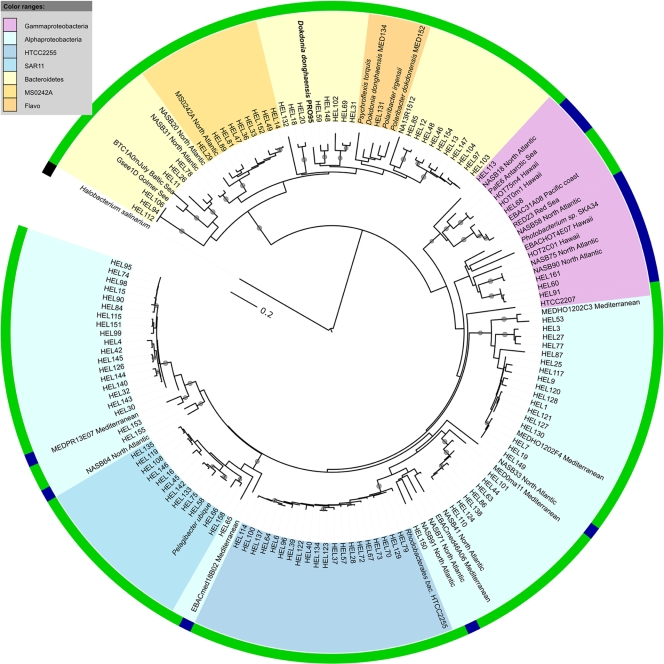
The obtained nucleotide sequences could be translated into 98 unique partial protein sequences. At a similarity level of 80%, as also applied in reference 6, 27 clusters of PR proteins could be identified (Fig. 2). Six of them contained more than eight sequences. Phylogenetic analyses revealed that 78 (66.7%) of the PR protein fragments in our analysis showed closest similarity to alphaproteobacterial PRs. The four largest clusters were found within this group. A cluster of 20 sequences (18% of all PRs) showed high degrees of similarity (>90% identity) to PR from the cultivated Roseobacter strain HTCC2255. Until now, this bacterium had not been shown to be dominant in any marine habitat. Sequences related to the SAR11 clade bacterium “Ca. Pelagibacter ubique” accounted for 11% of Helgoland PRs. Bacterial artificial chromosome (BAC) clones from the Mediterranean Sea were the closest relatives to the two remaining clusters in this group. Thirty-four (29.0%) of the PR fragments were related to PRs from the Bacteroidetes group. The two clusters within this group were relatively distant from PRs of the cultivated flavobacterial species. The PR of strain PRO95, which we isolated from the same water sample yielding the other PR sequences obtained and which we classified as D. donghaensis (see below), is located in the second Helgoland cluster but is distant from the PR of D. donghaensis MED134. The fraction of Bacteroidetes PR-like proteins found in the sample was relatively high compared to, e.g., that in a clone library of PRs from the Sargasso Sea, where they accounted for 10% of all PRs (6). Only five (4.3%) of the PR sequences obtained were related to gammaproteobacterial PRs. None of the sequences showed a close relationship to the PR of the cultivated Photobacterium sp. SKA34 or the BAC clone HOT2C01. The latter has been demonstrated to be highly abundant within a clone library from the Sargasso Sea (6). We could not find any sequences related to those from the cultivated gammaproteobacterium HTCC2207 of the coastal SAR92 clade, which is due to the inability of the primers used to amplify SAR92 PR sequences (37).
FIG. 2.
Neighbor-joining tree based on PR protein sequences isolated from North Sea surface water near the island of Helgoland showing the phylogenetic distribution of PRs in the North Sea. Bootstrap values (for 1,000 replicates) of >80% are indicated by filled circles at the branches. The scale bar shows the number of amino acid substitutions per site. For comparison, PR sequences from isolates and clones from the North Atlantic and Pacific Oceans and the Baltic, Mediterranean, Red, and Arctic Seas have been included. Clone and isolate names are colored according to the relationship of the corresponding PRs to PRs from Bacteroidetes, Alphaproteobacteria, and Gammaproteobacteria. Clusters of more than 8 sequences within these groups are highlighted. The outer ring shows the predicted absorption of green or blue light as inferred from the amino acid sequence. The GenBank accession numbers of the sequences used for the alignment are as follows: Helgoland (HEL) PR clones (HEL1 to HEL161), FJ560751 to FJ560867; Halobacterium salinarum, BAA01801.1; Polaribacter dokdonensis, ZP_01054176.1; Polaribacter irgensii, ZP_01117885.1; Psychroflexis torquis, ZP_01253360.1; D. donghaensis MED134, ZP_01049273; EBACHOT4E07 (SAR86 clade), AAT38609.1; HOT2C01, AAR05342.1; HOT75m4, AAK30179.1; HOT0m1, AAK30176.1; Photobacterium sp. SKA34, ZP_01161099.1; EBAC20E09 (SAR86 clade), AAS73014.1; EBAC31A08, AAG10475.1; PalE6, AAK30200.1; EBACmed18B02 (SAR11 clade), AAY82751.1; MEDeBAC46A06 (SAR11 clade), AAY82845.1; “Ca. Pelagibacter ubique,” AAZ21446.1; RED23, AAO21449.1; HTCC2207 (SAR92 clade), ABO88140.1; BTC1A0mJuly, ACB55057.1; Gsee1D, ACB55102.1; NASB75, ABU49527.1; NASB90, ABU49536.1; NASB58, ABU49514.1; NASB18, ABU49502.1; NASB64, ABU49517.1; NASB41, ABU49506.1; NASB71, ABU49523.1; NASB91, ABU49537.1; NASB33, ABU49505.1; NASB28, ABU49498.1; NASB31, ABU49455.1; NASB20, ABU49456.1; MS0242A, ABM91108.1; MedPR13e07, AAY68041.1; and PRO95, ACM89772.1. Flavo, flavobacteria.
PR gene abundance.
We used a quantitative PCR approach to estimate the abundance of different PR gene groups compared to the abundance of bacterial 16S rRNA genes. In previous studies, it was assumed that one marine bacterium harbors only 1 PR gene copy but on average 1.9 copies of the 16S rRNA gene (6, 41). This factor was also used in our study to normalize the ratio of PR gene abundance to 16S rRNA gene abundance. We investigated PR sequences from the SAR11, HTCC2255, MEDHO1202F4, flavobacterial, and MS0242A clusters for their abundance in the Helgoland DNA sample. The results are summarized in Table 2. In good accordance with the findings from the diversity study, the most abundant PR gene (13.2%) was an HTCC2255 PR gene-like sequence; 8.7% of the bacteria carried a PR gene related to the MEDHO1202F4 PR gene, and 5.3% of the bacteria carried a SAR11 PR gene-like sequence. The latter amount is at the lower limit for SAR11 PR gene-like sequences from North Atlantic sampling sites outside the Sargasso Sea (6). Sequences from the MS0242A cluster of Bacteroidetes PR genes (amplified with the Flavo-HEL primer pair [Flavo-H_f and Flavo-H_r]) were found in 3.6% of all bacteria. Only 1.0% harbored a PR gene related to genes in cultivated flavobacteria which could be amplified with the Flavo-NASB primer pair (Flavo_F2 and Flavo_R2). Previously, the results of quantitative PCR with the Flavo-NASB primers indicated that flavobacterial PRs related to D. donghaensis MED134 PR account for only 2% (on average) of PRs in the Sargasso Sea and 0.3% of PRs in other North Atlantic sites (6). In the previous study, 10% of PR sequences in a clone library were of flavobacterial origin, but Flavo-NASB primer-amplified PR sequences from the same sampling site accounted for only 4% of all PR sequences, indicating that the Flavo-NASB primers are not suitable for the detection of all flavobacterial PRs. Because we did not test all clusters of PRs found in the diversity study, a determination of the total abundance of PR-producing bacteria is not possible. But considering that the quantitative PCR study covered ∼40% of all clones found in our library, a rough estimate would be that around 50% of all bacteria at the sampling site must carry a PR gene. Since the clone library underestimated the diversity of PR genes in the sample, the true value can be expected to be even higher.
TABLE 2.
Abundance of PR sequence types relative to abundance of bacterial 16S rRNA genesa
| PR cluster | Relative abundance | SDb |
|---|---|---|
| SAR11 | 0.053 | 0.005 |
| HTCC2255 | 0.131 | 0.008 |
| MEDHO1202F4 | 0.087 | 0.005 |
| MedPR13e07 | ND | ND |
| MS0242A | 0.038 | 0.004 |
| Flavo-HEL | 0.036 | 0.003 |
| Flavo-NASB | 0.010 | 0.006 |
| Total | 0.345 |
Relative abundances in a sample of surface water from the North Sea were determined by quantitative PCR. ND, not determined.
SD, standard deviation of results for four technical replicas.
PR spectral tuning.
We further analyzed the spectral tuning of the cloned PR fragments. A single amino acid in position 105 (EBAC31A08 numbering) determines the wavelength of the absorbance maximum of PRs (25). PRs absorbing green light (wavelength, ∼525 nm) carry a hydrophobic amino acid, like leucine, methionine, alanine, or valine, at this position, whereas PRs absorbing blue light (wavelength, ∼490 nm) have glutamine. The majority of the cloned PR fragments (114 of 117; 97.4%) were for green-light-absorbing PRs; at amino acid position 105, either a leucine (in 78 of 117 sequences; 66.7%) or a methionine (in 36 of 117 sequences; 30.8%) was present. The distributions of these two isoforms among the phylogenetic clades were strictly discrete: all the Bacteroidetes PR-like sequences carried a methionine at this position, and almost all alphaproteobacterial PRs carried a leucine. Sequences for blue-light-absorbing PRs were found in only 3 (2.5%) of the 117 cloned PR fragments, and they were related to PR sequences from the gammaproteobacterial SAR86 clade (Fig. 2; see also File S1 in the supplemental material). Thus, putative green-light-absorbing PRs strongly dominated in the surface water of the North Sea. This finding is in agreement with data from other studies. In samples from the Mediterranean Sea, between 9 and 62% of all PRs were green light absorbing, and these PRs were found mainly in samples from the upper water layers, whereas blue-light-absorbing PRs prevailed in samples from greater depths and were the only variant present in Sargasso Sea samples (31). The Sorcerer II GOS expedition also found that green-light-absorbing PRs are abundant in temperate Atlantic waters and coastal regions (from outside Canadian waters to the Chesapeake Bay) and that the blue-light-absorbing PR variant is more abundant in warmer waters farther from land (29).
Biogeography of Helgoland PRs.
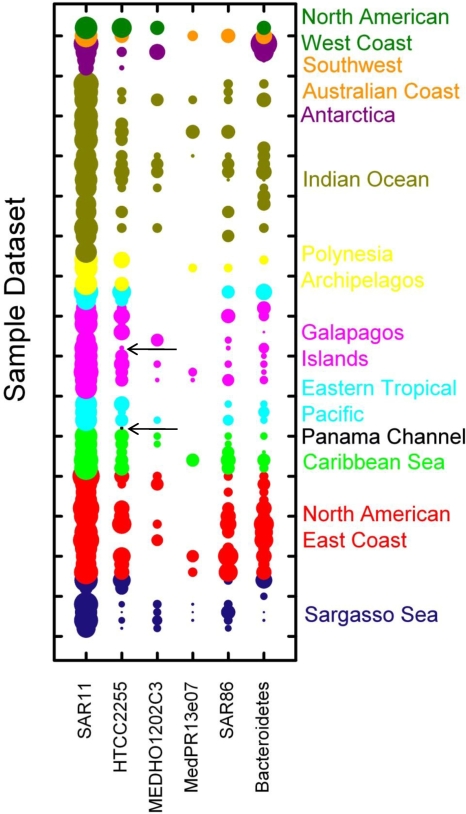
We scanned marine metagenomic databases obtained from the CAMERA website for sequences with high degrees of similarity to the PR genes of the Helgoland diversity study. Using BLAST with an E value exponent of −30 as the cutoff, we obtained 1,507 PR sequences from the GOS database, 74 PR sequences from the Antarctica aquatic data set, 18 PR sequences from the Botany Bay data set, and 10 PR sequences from the Monterey Bay data set. The abundance of PRs in total and in different groups and clusters was calculated as the number of PR fragments/Mb of sequence from the sampling site, and results ranged from the absence of PR sequences to 0.29 total PR fragments/Mb. PR abundance in coastal samples was significantly higher than that in open ocean samples (t = 2.3; P = 0.05). The collected data are summarized graphically in Fig. 3. It is interesting that the overall widespread Roseobacter HTCC2255 PR-like proteins are the only ones that are found in the freshwater of the Panama Channel and are the dominating PRs in a hypersaline lake (site GS033, Galapagos Islands), indicating that they may represent ecological generalists. PR sequences of the MedPR13e07 cluster were absent from almost all tested metagenomic samples, and those of the MEDHO1202F4 cluster were absent from at least half of the tested samples. Both clusters are highly abundant in the Helgoland sample. However, PR sequences related to these clusters as well as the HTCC2255 cluster were highly abundant in PCR-generated libraries from the Mediterranean Sea (31, 32), suggesting that a PCR bias may contribute to these differences.
FIG. 3.
Presence and relative abundance of Helgoland PR clusters in global metagenomic databases. Sampling sites represented in the GOS, Antarctica, Botany Bay, and Monterey Bay metagenomic databases are ordered and color coded by geographic location as indicated on the vertical axis. A detailed description of the sampling sites can be found at http://camera.calit2.net/. The relative abundance of PRs [log2(number of PR sequences/Gb)] from Helgoland clusters (horizontal axis) is visualized by the sizes of the circles. Note the data for the nonmarine Panama Channel and the hypersaline Galapagos Islands lake sample, where the only PR is the HTTC2255 PR-like protein (highlighted by arrows).
In summary, the levels of abundance and diversity of bacterial PRs in the shallow waters near the island of Helgoland are high. More than 50% of all bacteria are estimated to harbor a PR. HTCC2255-related and Bacteroidetes PR-like proteins dominate the community. The majority of the PRs can be expected to be green light absorbing based on the amino acid at signature position 105.
Cultivation of a strain producing a PR representative of PR clusters in the North Sea. (i) Screening for PR-producing strains.
Bacteria were isolated by plating of appropriate dilutions of the North Sea water sample onto plates containing only sterile North Sea water and agar. After 4 weeks of incubation, approximately 400 single colonies which were very small and colorless appeared. Colonies were restreaked onto marine agar for purification. All but two of the initial colonies were able to grow on full-strength marine agar. From the marine agar plates, single pure colonies were inoculated into liquid MB medium, and DNA was extracted. In parallel, glycerol stocks for each isolate were prepared. The genomic DNA was screened for PR sequences by PCR with degenerate primers (32) without applying selection criteria. No attempt was made to characterize the isolates phylogenetically. After 156 of the isolates had been tested, two PR-containing strains, PRO95 and PRO100, were discovered and further screening was stopped. Since the 16S rRNA gene sequences of these two strains were identical, all further analyses were carried out with strain PRO95.
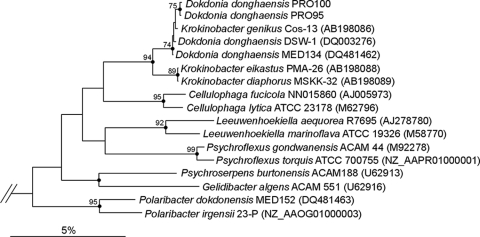
(ii) Phylogenetic affiliation of PRO95.
Phylogenetic analysis (Fig. 4) showed that PRO95 (and PRO100) was most closely affiliated with the type strains Krokinobacter genikus Cos-13 and D. donghaensis DSW-1. The species D. donghaensis was first described in 2005 by Yoon et al. (43), while K. genikus was described later (19) and would therefore probably have to be reclassified. Because of the high degree of sequence similarity (99.7%) between the 16S rRNA gene of PRO95 and that of D. donghaensis DSW-21, we tentatively assigned PRO95 the species identification D. donghaensis. The PR-containing strain PRO95 found in this study is a Gram-negative, aerobic, nonmotile bacterium which forms rods. Colonies on agar appear yellow (cream-colored to orange). These observations are in accordance with the description from Yoon et al. (43). The genus Dokdonia belongs to the class Flavobacteria and the phylum Bacteroidetes, a bacterial group which is abundant in marine environments (5, 16, 20). The 16S rRNA gene sequence of PRO95 is 99.7% identical to that of D. donghaensis DSW-21 (43) and 99.4% identical to that of D. donghaensis MED134, until now the only strain which was shown to grow better in the light than in the dark on extremely diluted medium (17).
FIG. 4.
Phylogenetic positions of strains PRO95 and PRO100 within the Flavobacteriaceae based on 16S rRNA gene comparisons. Phylogenetic analyses of 16S rRNA gene sequences were performed with the ARB software package (24). Only sequences longer than 1,400 bp were considered in the calculations. The tree was generated using the neighbor-joining method. Bootstrap values indicated at the nodes were derived from 1,000 replicates. Only values higher than 50% are shown. Filled circles indicate nodes also recovered reproducibly with maximum likelihood. Selected members of the Gammaproteobacteria were used as an out-group (data not shown). Bar, 0.05 substitutions per nucleotide position.
(iii) Complete PR gene of PRO95.
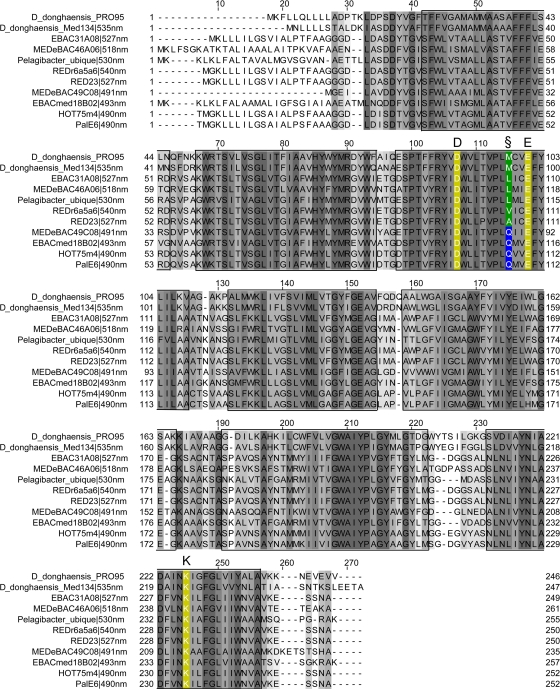
The amplified PR fragment from PRO95 was cloned and sequenced. The sequence of the complete PR gene was obtained by sequencing the genomic DNA with specific primers located inside the amplified fragment (Table 1). Finally, primers were designed for the complete gene, and it was amplified from the genomic DNA by PCR, cloned, and sequenced again for confirmation. The PR gene of D. donghaensis PRO95 encodes a protein of 246 amino acid residues. The amino acid sequence is 76% identical to those from D. donghaensis MED134 and P. dokdonensis MED152, two other flavobacteria which harbor PR (17, 18). The PR sequence of D. donghaensis PRO95 shows the typical features necessary for proton pumping, including Asp97 and Glu108 residues, which act as a proton acceptor and donor, respectively, in the retinylidene Schiff base transfer during the PR photocycle. The PR of PRO95 has a methionine at amino acid position 105, which is typical for green-light-absorbing PRs (Fig. 5).
FIG. 5.
Alignment of the PR of PRO95 with the PR of D. donghaensis MED134 and cloned PR sequences. Predicted transmembrane helices are marked by boxes. Gray shading indicates positions of conserved residues. Key amino acids for PR functionality (listed herein with EBAC31A08 numbering) are marked by colors: Lys131 (K) binds retinal, and Asp97 (D) and Glu108 (E) function as Schiff base proton acceptor and donor, respectively. Position 105 (§) plays a role in spectral tuning. Gln at position 105 (blue box) leads to absorption maxima at ∼490 nm. Met, Leu, Val, and Ala at this position (green box) result in absorption maxima at 518 to 535 nm. The GenBank accession numbers of the sequences used for the alignment are as follows: D. donghaensis MED134, ZP_01049273; EBAC31A08, AAG10475.1; MEDeBAC46A06 (SAR11 clade), AAY82845.1; “Ca. Pelagibacter ubique,” AAZ21446.1; REDr6a5a6, AAO21455.1; RED23, AAO21449.1; MEDeBAC49C08, AAY82659.1; EBACmed18B02 (SAR11 clade), AAY82751.1; HOT75m4, AAK30179.1; and PalE6, AAK30200.1.
(iv) The PRO95 PR gene falls into a Helgoland cluster of PR genes.
The cultivation-independent analysis of PR diversity in the North Sea reported above showed that in this location, the percentage of Bacteroidetes PR gene-related sequences (29%) is relatively high compared to that in the Sargasso Sea. These data also show that the PR gene of strain PRO95 falls into a cluster of PR genes found at the sampling site studied in this work (Fig. 2), which is distinct from the PR gene carried by flavobacterium strain MED134 from the Mediterranean Sea. Thus, the cultivated strain D. donghaensis PRO95 is representative of PR-producing flavobacteria in the North Sea. In a recent mesocosm study of the dynamics of bacterioplankton abundance and diversity during a phytoplankton bloom in a North Sea fjord 20 km south of Bergen, Norway, sequences related to PRO95 sequences were found among those with dominant denaturing gradient gel electrophoresis (DGGE) bands (8).
Effects of light and carbon source concentration on expression of PR in isolate PRO95. (i) Light does not stimulate growth of PRO95.
The growth of D. donghaensis PRO95 in diluted MB medium in a natural day-night cycle, in constant light, and in complete darkness was studied. The medium was supplemented with sea salts to keep the salinity constant, while the concentrations of the carbon sources peptone and yeast extract were decreased. No significant increase in the growth rate or the final OD in the light compared to that in the dark was found with the media tested, which corresponded to carbon concentrations between 9.7 and 121 mM (see File S2 in the supplemental material). Until now, only four isolates carrying the PR gene had been tested for an effect of light on growth (12). Only one of these isolates, D. donghaensis MED134, showed increased growth in the light at carbon concentrations between 0.1 and 1.1 mM but not in full-strength medium (17), whereas all other isolates grew equally well in the light and in the dark. The carbon sources used in the previous study (17) were the same as those in our study, i.e., peptone and yeast extract in a ratio of 5:1, corresponding to 242 mM carbon in full-strength medium. A small increase in cell density may not have been detectable by the OD measurements used in our study. Therefore, it remains to be investigated if more sensitive cell density-related parameters, e.g., direct microscopic cell counts, would reveal a stimulatory effect of light on the growth of PRO95, especially at nutrient concentrations below 9.7 mM carbon, corresponding to 200 mg peptone and 40 mg yeast extract per liter of seawater. Alternatively, the cellular energy presumably generated by PR in the light may be used for the synthesis of storage compounds.
(ii) The PR gene is expressed constitutively in PRO95.
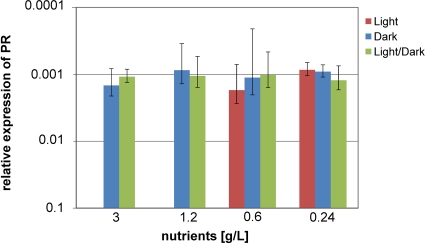
The expression of the PR gene in D. donghaensis PRO95 was investigated by extraction of the mRNA, reverse transcription, and amplification of a fragment of the PR gene from the cDNA with specific primers (Table 1) by quantitative PCR (qRT-PCR). As a reference for quantification, the 16S rRNA gene was used. These experiments were carried out with up to four independent biological replicas of cultures grown in the light, in the dark, and in a natural light-dark cycle in MB preparations diluted from 1/2 down to 1/25 (corresponding to 121 mM C to 9.7 mM C [see File S2 in the supplemental material]). Figure 6 shows that PR was expressed to similar extents under all conditions. Neither high nutrient concentrations nor constant darkness repressed the expression of PR. To the best of our knowledge, the expression of PR by a flavobacterium cultivated at these carbon concentrations has not been quantified previously.
FIG. 6.
Effects of carbon concentration and light on PR expression in PRO95 as determined by qRT-PCR. Bacteria were cultivated under the conditions indicated. Nutrients were peptone and yeast extract in the ratio 5:1. RNA was extracted, cDNA was generated, and the abundance of PR transcripts in relation to that of the 16S rRNA gene was determined (see Materials and Methods for details). Means and standard deviations for two to four independent biological replicas are shown. The standard error for technical replicas was ∼1% (data not shown).
The data are in contrast to those from a recent mesocosm study of intact microbial communities in the ocean, which found that light increased both the expression of PR and the abundance of PR-producing bacteria (21). However, although these data strongly suggest that light stimulated the growth of PR-containing bacteria, the possibility that this stimulation was an indirect effect caused, e.g., by changes in the population of phototrophic microalgae, grazing by protozoa, or viral mortality cannot be totally excluded.
The constitutive expression of PR in PRO95 may be the result of the loss of regulatory functions as an adaptation to life on rich laboratory medium. The North Sea isolates obtained in this study, including PRO95, grew originally on nutrient-poor medium at 16°C for 4 weeks, forming very tiny colonies. However, all but two of them could be restreaked onto full-strength marine agar, where they grew well at room temperature. It is not known if irreversible genetic changes occur during these adaptation processes, e.g., by selection for certain clones, or if regulatory circuits are turned off only transiently. In the latter case, regulated expression of PR may be detectable at carbon concentrations below 9.7 mM. Alternatively, the data may indicate that PR is expressed constitutively because the advantage of using light as a supplementary energy source outweighs the costs of expressing PR even in the absence of light.
Conclusions.
PRs in a near-surface water sample from the long-term marine monitoring station Kabeltonne in the North Sea are highly abundant and diverse. At least 50% of all bacteria in the sample harbor PR. The majority of the PRs in the North Sea bacteria are green light absorbing, based on the signature amino acid at position 105. The sampling site is characterized by the relatively high abundances of Bacteroidetes and Roseobacter HTCC2255 PR-like sequences. Two PR-containing flavobacteria were isolated from this water sample by using oligocarbophilic media. The strains are representative of a PR-producing cluster of flavobacteria in the North Sea. The strain studied in detail, D. donghaensis PRO95, expressed the PR gene in medium diluted to a carbon concentration of 9.7 mM, both in the light and in the dark. Light did not stimulate growth. Possibly, more sensitive methods will be required to detect the physiological function of PR in PRO95.
Supplementary Material
Acknowledgments
We are grateful to the Alfred Wegener Institute for Polar Research (AWI) Biological Station of Helgoland, Germany, especially Markus Molis, Andreas Wagner, Margret Krüss, Brigitte Rauch, Ulf Bickmeyer, and Christian Schütt, for providing facilities for guest researchers and for the support during lab courses. We thank Helmut Blöcker and his group (HZI) for sequencing our samples. Finally, we thank Helena Sztajer for her kind assistance.
Thanks to Dieter Jahn, Technical University of Braunschweig, for financial and logistic support of our lab courses at Helgoland.
Footnotes
Published ahead of print on 19 March 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Atamna-Ismaeel, N., G. Sabehi, I. Sharon, K. P. Witzel, M. Labrenz, K. Jurgens, T. Barkay, M. Stomp, J. Huisman, and O. Beja. 2008. Widespread distribution of proteorhodopsins in freshwater and brackish ecosystems. ISME J. 2:656-662. [DOI] [PubMed] [Google Scholar]
- 3.Beja, O., L. Aravind, E. V. Koonin, M. T. Suzuki, A. Hadd, L. P. Nguyen, S. B. Jovanovich, C. M. Gates, R. A. Feldman, J. L. Spudich, E. N. Spudich, and E. F. DeLong. 2000. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289:1902-1906. [DOI] [PubMed] [Google Scholar]
- 4.Beja, O., E. N. Spudich, J. L. Spudich, M. Leclerc, and E. F. DeLong. 2001. Proteorhodopsin phototrophy in the ocean. Nature 411:786-789. [DOI] [PubMed] [Google Scholar]
- 5.Bowman, J. P., S. A. McCammon, M. V. Brown, D. S. Nichols, and T. A. McMeekin. 1997. Diversity and association of psychrophilic bacteria in Antarctic sea ice. Appl. Environ. Microbiol. 63:3068-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, B. J., L. A. Waidner, M. T. Cottrell, and D. L. Kirchman. 2008. Abundant proteorhodopsin genes in the North Atlantic Ocean. Environ. Microbiol. 10:99-109. [DOI] [PubMed] [Google Scholar]
- 7.Chao, A. 1984. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 11:265-270. [Google Scholar]
- 8.Cunliffe, M., A. S. Whiteley, L. Newbold, A. Oliver, H. Schafer, and J. C. Murrell. 2009. Comparison of bacterioneuston and bacterioplankton dynamics during a phytoplankton bloom in a fjord mesocosm. Appl. Environ. Microbiol. 75:7173-7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de la Torre, J. R., L. M. Christianson, O. Beja, M. T. Suzuki, D. M. Karl, J. Heidelberg, and E. F. DeLong. 2003. Proteorhodopsin genes are distributed among divergent marine bacterial taxa. Proc. Natl. Acad. Sci. U. S. A. 100:12830-12835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franke, H.-D., F. Buchholz, and K. H. Wiltshire. 2004. Ecological long-term research at Helgoland (German Bight, North Sea): retrospect and prospect—an introduction. Helgol. Mar. Res. 58:223-229. [Google Scholar]
- 11.Frigaard, N. U., A. Martinez, T. J. Mincer, and E. F. DeLong. 2006. Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. Nature 439:847-850. [DOI] [PubMed] [Google Scholar]
- 12.Fuhrman, J. A., M. S. Schwalbach, and U. Stingl. 2008. Proteorhodopsins: an array of physiological roles? Nat. Rev. Microbiol. 6:488-494. [DOI] [PubMed] [Google Scholar]
- 13.Gerdts, G., A. Wichels, H. Döpke, K.-W. Klings, W. Gunkel, and C. Schütt. 2004. 40-year long-term study of microbial parameters near Helgoland (German Bight, North Sea): historical view and future perspectives. Helgol. Mar. Res. 58:230-242. [Google Scholar]
- 14.Giovannoni, S. J., L. Bibbs, J. C. Cho, M. D. Stapels, R. Desiderio, K. L. Vergin, M. S. Rappe, S. Laney, L. J. Wilhelm, H. J. Tripp, E. J. Mathur, and D. F. Barofsky. 2005. Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature 438:82-85. [DOI] [PubMed] [Google Scholar]
- 15.Giovannoni, S. J., D. H. Hayakawa, H. J. Tripp, U. Stingl, S. A. Givan, J. C. Cho, H. M. Oh, J. B. Kitner, K. L. Vergin, and M. S. Rappe. 2008. The small genome of an abundant coastal ocean methylotroph. Environ. Microbiol. 10:1771-1782. [DOI] [PubMed] [Google Scholar]
- 16.Glockner, F. O., B. M. Fuchs, and R. Amann. 1999. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65:3721-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomez-Consarnau, L., J. M. Gonzalez, M. Coll-Llado, P. Gourdon, T. Pascher, R. Neutze, C. Pedros-Alio, and J. Pinhassi. 2007. Light stimulates growth of proteorhodopsin-containing marine Flavobacteria. Nature 445:210-213. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez, J. M., B. Fernandez-Gomez, A. Fernandez-Guerra, L. Gomez-Consarnau, O. Sanchez, M. Coll-Llado, J. Del Campo, L. Escudero, R. Rodriguez-Martinez, L. Alonso-Saez, M. Latasa, I. Paulsen, O. Nedashkovskaya, I. Lekunberri, J. Pinhassi, and C. Pedros-Alio. 2008. Genome analysis of the proteorhodopsin-containing marine bacterium Polaribacter sp. MED152 (Flavobacteria). Proc. Natl. Acad. Sci. U. S. A. 105:8724-8729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan, S. T., Y. Nakagawa, and S. Harayama. 2006. Krokinobacter gen. nov., with three novel species, in the family Flavobacteriaceae. Int. J. Syst. Evol. Microbiol. 56:323-328. [DOI] [PubMed] [Google Scholar]
- 20.Kirchman, D. L. 2002. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol. Ecol. 39:91-100. [DOI] [PubMed] [Google Scholar]
- 21.Lami, R., M. T. Cottrell, B. J. Campbell, and D. L. Kirchman. 2009. Light-dependent growth and proteorhodopsin expression by Flavobacteria and SAR11 in experiments with Delaware coastal waters. Environ. Microbiol. 11:3201-3209. [DOI] [PubMed] [Google Scholar]
- 22.Lane, D. J. 1991. 16S-23S rRNA sequencing, p. 125-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, Chichester, United Kingdom.
- 23.Letunic, I., and P. Bork. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127-128. [DOI] [PubMed] [Google Scholar]
- 24.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Man, D., W. Wang, G. Sabehi, L. Aravind, A. F. Post, R. Massana, E. N. Spudich, J. L. Spudich, and O. Beja. 2003. Diversification and spectral tuning in marine proteorhodopsins. EMBO J. 22:1725-1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez, A., A. S. Bradley, J. R. Waldbauer, R. E. Summons, and E. F. DeLong. 2007. Proteorhodopsin photosystem gene expression enables photophosphorylation in a heterologous host. Proc. Natl. Acad. Sci. U. S. A. 104:5590-5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCarren, J., and E. F. DeLong. 2007. Proteorhodopsin photosystem gene clusters exhibit co-evolutionary trends and shared ancestry among diverse marine microbial phyla. Environ. Microbiol. 9:846-858. [DOI] [PubMed] [Google Scholar]
- 28.Oesterhelt, D., and W. Stoeckenius. 1971. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat. New Biol. 233:149-152. [DOI] [PubMed] [Google Scholar]
- 29.Rusch, D. B., A. L. Halpern, G. Sutton, K. B. Heidelberg, S. Williamson, S. Yooseph, D. Wu, J. A. Eisen, J. M. Hoffman, K. Remington, K. Beeson, B. Tran, H. Smith, H. Baden-Tillson, C. Stewart, J. Thorpe, J. Freeman, C. Andrews-Pfannkoch, J. E. Venter, K. Li, S. Kravitz, J. F. Heidelberg, T. Utterback, Y. H. Rogers, L. I. Falcon, V. Souza, G. Bonilla-Rosso, L. E. Eguiarte, D. M. Karl, S. Sathyendranath, T. Platt, E. Bermingham, V. Gallardo, G. Tamayo-Castillo, M. R. Ferrari, R. L. Strausberg, K. Nealson, R. Friedman, M. Frazier, and J. C. Venter. 2007. The Sorcerer II Global Ocean Sampling Expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 5:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sabehi, G., O. Beja, M. T. Suzuki, C. M. Preston, and E. F. DeLong. 2004. Different SAR86 subgroups harbour divergent proteorhodopsins. Environ. Microbiol. 6:903-910. [DOI] [PubMed] [Google Scholar]
- 31.Sabehi, G., B. C. Kirkup, M. Rozenberg, N. Stambler, M. F. Polz, and O. Beja. 2007. Adaptation and spectral tuning in divergent marine proteorhodopsins from the eastern Mediterranean and the Sargasso Sea. ISME J. 1:48-55. [DOI] [PubMed] [Google Scholar]
- 32.Sabehi, G., A. Loy, K. H. Jung, R. Partha, J. L. Spudich, T. Isaacson, J. Hirschberg, M. Wagner, and O. Beja. 2005. New insights into metabolic properties of marine bacteria encoding proteorhodopsins. PLoS Biol. 3:e273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sabehi, G., R. Massana, J. P. Bielawski, M. Rosenberg, E. F. DeLong, and O. Beja. 2003. Novel proteorhodopsin variants from the Mediterranean and Red Seas. Environ. Microbiol. 5:842-849. [DOI] [PubMed] [Google Scholar]
- 34.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 35.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spudich, J. L., C. S. Yang, K. H. Jung, and E. N. Spudich. 2000. Retinylidene proteins: structures and functions from archaea to humans. Annu. Rev. Cell Dev. Biol. 16:365-392. [DOI] [PubMed] [Google Scholar]
- 37.Stingl, U., R. A. Desiderio, J. C. Cho, K. L. Vergin, and S. J. Giovannoni. 2007. The SAR92 clade: an abundant coastal clade of culturable marine bacteria possessing proteorhodopsin. Appl. Environ. Microbiol. 73:2290-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki, M. T., L. T. Taylor, and E. F. DeLong. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 66:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 40.Thompson, J. D., T. J. Gibson, and D. G. Higgins. 2003. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics 2003:2.3.1-2.3.22. [DOI] [PubMed] [Google Scholar]
- 41.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 42.Walter, J. M., D. Greenfield, C. Bustamante, and J. Liphardt. 2007. Light-powering Escherichia coli with proteorhodopsin. Proc. Natl. Acad. Sci. U. S. A. 104:2408-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoon, J. H., S. J. Kang, C. H. Lee, and T. K. Oh. 2005. Dokdonia donghaensis gen. nov., sp. nov., isolated from sea water. Int. J. Syst. Evol. Microbiol. 55:2323-2328. [DOI] [PubMed] [Google Scholar]
- 44.Zeng, Y. H., X. H. Chen, and N. Z. Jiao. 2007. Genetic diversity assessment of anoxygenic photosynthetic bacteria by distance-based grouping analysis of pufM sequences. Lett. Appl. Microbiol. 45:639-645. [DOI] [PubMed] [Google Scholar]
- 45.Zhao, M., F. Chen, and N. Jiao. 2008. Genetic diversity and abundance of flavobacterial proteorhodopsin in China seas. Appl. Environ. Microbiol. 75:529-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zuckerkandl, E., and L. Pauling. 1965. Molecules as documents of evolutionary history. J. Theor. Biol. 8:357-366. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.