Abstract
Preventing the formation of insoluble polyglutamine containing protein aggregates in neurons may represent an attractive therapeutic strategy to ameliorate Huntington's disease (HD). Therefore, the ability to screen for small molecules that suppress the self-assembly of huntingtin would have potential clinical and significant research applications. We have developed an automated filter retardation assay for the rapid identification of chemical compounds that prevent HD exon 1 protein aggregation in vitro. Using this method, a total of 25 benzothiazole derivatives that inhibit huntingtin fibrillogenesis in a dose-dependent manner were discovered from a library of ≈184,000 small molecules. The results obtained by the filter assay were confirmed by immunoblotting, electron microscopy, and mass spectrometry. Furthermore, cell culture studies revealed that 2-amino-4,7-dimethyl-benzothiazol-6-ol, a chemical compound similar to riluzole, significantly inhibits HD exon 1 aggregation in vivo. These findings may provide the basis for a new therapeutic approach to prevent the accumulation of insoluble protein aggregates in Huntington's disease and related glutamine repeat disorders.
Huntington's disease (HD) is a progressive neurodegenerative disorder with no effective treatment (1). The disease is caused by an elongated CAG trinucleotide repeat expansion located within exon 1 of the IT-15 gene encoding huntingtin, an ≈350-kDa protein of unknown function. The CAG repeat is translated into a polyglutamine (polyQ) stretch. In HD patients, huntingtin is expressed with 38–180 glutamine residues, whereas in healthy individuals the protein is synthesized with 8–37 glutamine residues (2, 3). Thus, the disease develops when a critical length of about 37 glutamine residues (pathological threshold) is exceeded, whereas a polyQ tract of fewer than 37 glutamine residues is tolerated by neuronal cells (4).
The accumulation of ubiquitinated polyQ-containing protein aggregates in neuronal inclusions is a pathological hallmark of HD and related glutamine repeat disorders (5). Whether the formation of huntingtin aggregates in brain is the cause, or merely the consequence, of disease, however, is still unclear (6). Within the last few years, aggregation of polyQ-containing proteins has been reproduced in various in vitro and in vivo model systems (7–11). Evidence has been presented that the process of aggregate formation is causally linked with disease progression. Recently, Yamamoto et al. (12) have shown that blockage of HD exon 1 expression in symptomatic transgenic mice results in disappearance of insoluble protein aggregates, as well as motor dysfunction, suggesting that protein aggregation in vivo is associated with disease progression.
The formation of insoluble polyQ-containing protein aggregates in vitro and in cell culture model systems has been inhibited by specific antibodies (13), peptides (14), heat shock proteins (15, 16), and chemical compounds (17). Moreover, antibodies (18), β-sheet breaker peptides (19), and chemical compounds (20–24) that prevent protein aggregation in various models of Alzheimer's and prion diseases have been described. However, the effect of these and other potential therapeutic molecules on the progression of neurodegenerative disorders in humans needs to be evaluated.
In this study, we have developed an automated filter retardation assay for the identification of HD exon 1 aggregation inhibitors. In contrast to conventional methods, our assay is eminently suited for high-throughput detection of chemical compounds that block huntingtin fibrillogenesis because it allows the parallel, nonradioactive screening of up to 384 potential aggregation inhibitors on a single filter membrane. Here, we report the rapid screening and characterization of a previously uncharacterized class of polyglutamine aggregation inhibitors from large chemical compound libraries by using this approach.
Materials and Methods
Chemical Compounds, Enzymes, and Instrumentation.
A library containing 184,880 chemical compounds was provided by Merck. All compounds were dissolved in 100% DMSO at a conc. of 3 mM. Riluzole was purchased from BioTrend (Cologne, Germany). Doxycycline and elastase were obtained from Sigma-Aldrich, and trypsin (modified version) was purchased from La Roche (Mannheim, Germany). The Cy3-labeled donkey anti-rabbit IgG was obtained from Jackson ImmunoResearch. Parallel liquid handling in 96-and 384-well format plates was performed with Tomtec (Orange, CT) Quadra 96SV and Quadra 384S robots.
Construction of Plasmids and Protein Purification.
Standard protocols for DNA manipulations were followed. Escherichia coli SURE (stop unwanted rearrangement events; Stratagene) was used as host strain for plasmid construction and protein expression. Plasmids pCAG51 and pCAG51ΔP are described elsewhere (7, 25). E. coli SURE carrying the plasmids pCAG51 or pCAG51ΔP was used for the expression of GST-HD51 (GST-HD exon 1 fusion protein with 51 glutamines) and GST-HD51ΔP fusion proteins, respectively. Recombinant proteins were purified under native conditions by affinity chromatography on glutathione agarose as described (25), and aliquots were subjected to matrix-assisted laser desorption ionization–time-of-flight (MALDI-TOF) MS for sequence confirmation. Recombinant proteins were stored at a conc. of 25 μM at −80°C.
High-Throughput Screening Procedure.
In each well of six hundred 384-well microtiter plates, 15 μl of a 20 μM solution of the chemical compound to be tested (in 150 mM NaCl, 20 mM Tris⋅HCl (pH 8.0), 2 mM CaCl2, and 6.7% DMSO) was mixed with 15 μl of predigested GST-HD51 fusion protein (1.25 μM) by using a pipetting robot. Total removal of the GST tag from the fusion protein GST-HD51 was achieved by elastase treatment (3 min at 37°C) before the addition of chemical compounds. Samples were incubated for 16 h at 37°C to allow aggregate formation. Reactions were stopped by addition of 30 μl of 4% SDS/100 mM DTT, followed by heating at 98°C for 10 min. Aliquots corresponding to 250 ng of GST-HD51 protein were filtered through a cellulose acetate membrane (0.2 μm, Schleicher & Schuell) by using a 384-well vacuum dot blot apparatus. Captured aggregates were detected by incubation with HD1 antibody (diluted 1:5,000), followed by incubation with alkaline phosphatase-conjugated anti-rabbit secondary antibody and the fluorescent substrate AttoPhos. Signals corresponding to SDS-insoluble aggregates were quantified by using aida 2.0 image analysis software (Raytest, Straubenhardt, Germany).
SDS/PAGE and Western Blotting.
Proteins present in whole cell extracts or aggregation reactions were denatured, separated by SDS/PAGE (10 or 12%), and transferred to nitrocellulose. Membranes were blocked with 3% nonfat dry milk in Tris-buffered saline (TBS) containing 0.05% Tween 20 and incubated with the HD1 antibody (7). Secondary antibody was peroxidase-conjugated anti-rabbit IgG (Roche). Immunoreactive protein was detected by using enhanced chemiluminescence reagent (ECL, Amersham Pharmacia).
MS and Microscopic Analysis.
The conditions for proteolytic digestion of GST-HD51ΔP with trypsin have been described (17). Mass maps of digested proteins were recorded on a Bruker Scout MTP Reflex III MALDI mass spectrometer (Bruker Daltonik, Germany) using the matrix α-cyano-4-hydroxycinamic acid. For electron microscopic analysis, the trypsin-digested GST-HD51 fusion protein was adjusted to a final conc. of 50 μg/ml in 40 mM Tris⋅HCl (pH 8.0) and 150 mM NaCl. Samples were negatively stained with 1% uranyl acetate and viewed in a Philips CM100 electron microscope (Philips Electron Optics, Eindhoven, The Netherlands). Electron micrographs of perinuclear inclusion bodies in 293 Tet-Off cells were generated as described (11).
Culturing of 293 Tet-Off Cells.
Cells were grown in l-glutamine-free minimum essential medium with Earle's salts (GIBCO/BRL) supplemented with 10% FBS, 2 mM l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, 100 μg/ml G418, 150 μg/ml hygromycin B, and 10 ng/ml doxycycline in poly-l-lysine-coated cell culture flasks. HDQ51 (Flag-tagged HD exon 1 protein with 51 glutamines) expression was induced by thoroughly washing the cells with PBS and adding fresh medium lacking doxycycline. After incubation for 4 h in the absence of doxycycline, chemical compounds were added to the medium at the indicated concentrations. The compound-containing medium was changed daily, and, after 72–76 h of incubation, cells were harvested and protein extracts were prepared as described (26). Aliquots corresponding to 10–25 μg of protein were used for the cellulose acetate filter retardation assay (25). Protein concentration was determined by using the Bradford protein assay.
Indirect Immunofluorescence Microscopy.
293 Tet-Off cells were grown in Leighton tubes (Costar) in the presence or absence of PGL-135 (conc. 25 or 50 μM). Expression of HDQ51 was induced for 3 days by removal of doxycycline from the culture medium. Chemical compounds were added 4 h after incubation of cells in doxycycline-free medium. Indirect immunofluorescence microscopy was performed after 3 days as described (11).
Results
Inhibitor Screening.
To identify polyQ aggregation inhibitors from large chemical libraries, a high-throughput in vitro screening assay was developed. The principle of this automated filter retardation assay is shown schematically in Fig. 1A. The assay is based on the finding that polyQ-containing protein aggregates are resistant to SDS and selectively retained on a cellulose acetate filter, whereas SDS-soluble protein under the same conditions is not. The aggregates retained on the filter membrane are then detected and quantified by immunoblot analysis using specific antibodies (25).
Figure 1.
Development of an automated filter retardation assay for high-throughput drug screening. (A) Flow chart of the automated filter retardation assay. (B) Effect of chemical compounds on HD exon 1 aggregation as monitored by the filter assay. GST-HD51 fusion protein at a conc. of 1.25 μM was predigested for 3 min at 37°C with elastase to remove totally the GST tag from the fusion protein and to initiate aggregation of the HDQ51 protein. Immediately after this step, various chemical compounds were added (final conc. 10 μM) and samples were incubated for an additional 16 h at 37°C. Then, aggregation reactions were filtered through a cellulose acetate membrane by using a 384-well dot blot apparatus. Captured aggregates were detected by immunoblotting using the HD1 antibody. On each filter membrane, 320 different chemical compounds were tested (squares A2–H11). Squares A1–H1 and A12–H12 were used for control samples. In square F8, an inhibitory compound was identified. ThioS, thioflavine S.
GST-HD51 was incubated for 3 min at 37°C with elastase, resulting in the complete removal of the GST tag from the fusion protein and the initiation of the aggregation process (data not shown). Then, immediately, chemical compounds were added to give a final conc. of 10 μM by using a pipetting robot. Incubation was continued for 16 h at 37°C to permit aggregate formation. Proteins were reduced and denatured by boiling in 2% SDS/50 mM DTT and filtered through a cellulose acetate membrane by using a 384-well dot blot apparatus. After washing of the membrane with 0.2% SDS, the amount of insoluble polyQ-containing protein aggregates retained on the surface was quantified by immunoblotting and subsequent image analysis. Compounds that reduced the signal intensity by more than 15% relative to the signal intensity of noninhibited HD51 aggregation reactions were identified as hits (Fig. 1B, F8). As positive controls, elastase-digested GST-HD51 protein treated with the known polyglutamine aggregation inhibitors Congo red and thioflavine S (17) (control I), as well as undigested GST-HD51 protein (control II), which does not aggregate (7), were applied to the filter membrane. Samples treated with the solvent DMSO and untreated GST-HD51 elastase cleavage products were used as negative controls (Fig. 1B).
High-throughput screening of an ≈184,000-chemical compound library by using the automated filter retardation assay resulted in the identification of about 300 chemical compounds that inhibited HD51 protein aggregation in a dose-dependent manner. Among these, we identified 25 benzothiazole derivatives by cluster analysis. Benzothiazole derivatives have been shown previously to be effective in treating neurodegenerative disorders such as amyotrophic lateral sclerosis (27). For example, riluzole (2-amino-6-trifluoromethoxybenzothiazole), a potent antagonist of glutamate release, has been reported to slow down disease progression in amyotrophic lateral sclerosis patients (28, 29). First clinical data on riluzole treatment of HD patients indicate a positive effect on hyperkinesia (30), and recently riluzole was reported to prolong the lifespan of a mouse model of HD (31). However, its mode of action is largely unknown. The identification of benzothiazoles as polyglutamine aggregation inhibitors is highly relevant because they are very promising candidates for future drug development. The structures of the eight most effective compounds in the in vitro screen having IC50 values in the range of 1–11 μM are shown in Table 1.
Table 1.
Benzothiazoles inhibit HD exon 1 aggregation in vitro
IC50 values shown are the average of four independent determinations (±SE).
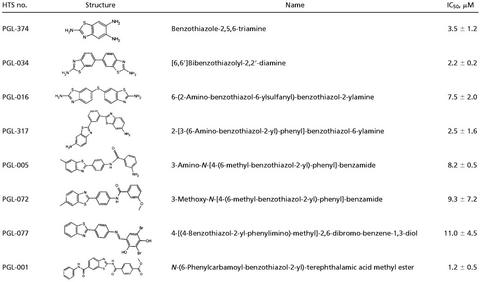
The effect of increasing concentrations of the compounds PGL-001, PGL-005, and PGL-034 on HD51 aggregation as monitored by the filter retardation assay is shown in Fig. 2 A and B. PGL-001, PGL-005, and PGL-034 inhibited HD51 aggregation with IC50 values of 1.2, 8.2, and 2.2 μM, respectively, whereas the solvent DMSO alone was ineffective. The inhibitory effect of the benzothiazole derivatives PGL-001 and PGL-034 on HD51 aggregation in vitro was also confirmed by electron microscopy (Fig. 3A). After an 18-h incubation of trypsin-digested GST-HD51 fusion protein (final conc. 100 ng/μl) without added chemical compound, numerous clusters of high- molecular-weight fibrils with a ribbon-like morphology (diameter, 20–40 nm) were detected. In strong contrast, treatment of the trypsin-digested GST-HD51 protein with PGL-001 or PGL-034 (each at 20 μM) resulted in the appearance of only a few fibrils with a diameter of 6–8 nm. In agreement with previous observations (7), electron microscopy of undigested GST-HD51 protein (negative control) predominantly showed spherical particles with a diameter of 6–7 nm.
Figure 2.
In vitro inhibition of HD exon 1 aggregation by benzothiazoles. (A) Effect of the indicated chemical compounds on HD exon 1 aggregation as monitored by the filter retardation assay. (B) Quantification of the dot blot results shown in A. The signal intensity from the sample without added chemical compound (solvent) was arbitrarily set as 100. For structure of the chemical compounds, see Table 1.
Figure 3.
Analysis of HD exon 1 aggregation in vitro by using secondary assays. (A) Electron micrographs of HD51 fibrils formed in the presence or absence of the indicated benzothiazoles. Trypsin-digested GST-HD51 protein at 1.5 μM was incubated for 16 h at 37°C either without chemical compound (pos. control) or with PGL-001 and PGL-034 (final conc. 20 μM). Representative examples of fibrillar structures are shown. (B) Western blot analysis of trypsin-digested GST-HD51 fusion protein. Aggregation reactions were performed for 16 h in the presence or absence of the indicated chemical compounds. Where indicated (+), aliquots were taken and incubated for additional 30 min with 10 ng/μl proteinase K. Samples corresponding to 200 ng of fusion protein were analyzed by SDS/PAGE and immunoblotting using the HD1 antibody. (C) Effect of the compound PGL-001 on HD exon 1 aggregation as monitored by MALDI-TOF MS. GST-HD51ΔP at 2.5 μM was incubated for 18 h at 37°C with trypsin in the absence (control) or presence of PGL-001 (final conc. 20 μM). Analysis by MALDI-TOF MS revealed that, only in the presence of PGL-001, monomeric HD51ΔP peptide (monoisotopic mass of 8,074.74 Da) is detectable; without added compound, the peptide has assembled into insoluble high-molecular-weight protein aggregates that cannot be detected by MALDI-TOF MS.
Recently, we have shown that formation of insoluble HD exon 1 protein aggregates in vitro is a nucleation-dependent process (32) and that addition of chemical compounds such as Congo red or thioflavine S to the aggregation reaction significantly delays the assembly of monomeric HD exon 1 protein into high-molecular-weight fibrillar structures (17). To test whether the benzothiazole derivatives PGL-001 and PGL-034 have a similar effect on HD51 protein aggregation, a time course experiment was performed in which the decrease of soluble HD51 protein in the presence or absence of chemical compounds was monitored by SDS/PAGE and immunoblotting (Fig. 3B). After 24 h of incubation of trypsin-digested GST-HD51 protein, SDS-soluble HD51 protein was detected only in the presence of PGL-001 and PGL-034, indicating that these compounds have delayed the assembly of monomeric HD51 into insoluble protein aggregates. The soluble HD51 protein that resisted aggregate formation was readily accessible to proteolytic degradation. Thus, incubation for 30 min with proteinase K resulted in complete degradation of HD51 protein, whereas the high- molecular-weight polyQ-containing protein aggregates were largely resistant to proteinase K treatment (Fig. 3B). The results obtained by SDS/PAGE and immunoblotting were confirmed by MALDI-TOF MS analyses using a GST-HD exon 1 fusion protein, GST-HD51ΔP, that lacks the proline-rich region C-terminal to the polyQ tract (25). As shown in Fig. 3C, after incubation for 24 h, soluble HD51ΔP protein was detected only in samples that contained the compound PGL-001. In control samples lacking an added chemical compound, HD51ΔP assembled quantitatively into high-molecular-weight aggregates that are not accessible to MALDI-TOF MS analysis (17). Similar results were obtained with PGL-005 and PGL-034 (data not shown), indicating that benzothiazoles efficiently slow down the assembly of HD exon 1 protein into protease-resistant high-molecular-weight aggregates in vitro.
Inhibition of HD Exon 1 Protein Aggregation in a Cell Culture Model System of HD.
To determine whether benzothiazoles are also capable of inhibiting HD exon 1 protein aggregation in vivo, a cell-based 96-well microtiter plate huntingtin aggregation assay was developed. In this assay, the tetracycline (tet)-regulated expression system (33) was used for the synthesis of HDQ51 in 293 Tet-Off cells (11). Cultivation of cells for 48–72 h in the absence of doxycycline induces the expression of HDQ51, resulting in the formation of large perinuclear inclusion bodies that mainly consist of fibrillar HDQ51 protein (11). The HDQ51 fibrils formed in 293 Tet-Off cells have a diameter of ≈10 nm (data not shown).
Induced 293 Tet-Off cells expressing HDQ51 were grown for 72 h in the presence or absence of benzothiazole derivatives. Then, the cells were lysed, and the protein concentration in the cell extracts was determined. Because the protein concentration largely correlates with the cell density, a reduction of protein concentration by the addition of a given chemical compound indicates that the compound is toxic for 293 Tet-Off cells. For the detection of SDS-insoluble protein aggregates, protein extracts were filtered through a cellulose acetate membrane, and the amount of captured aggregates was quantified by immunoblotting and image analysis.
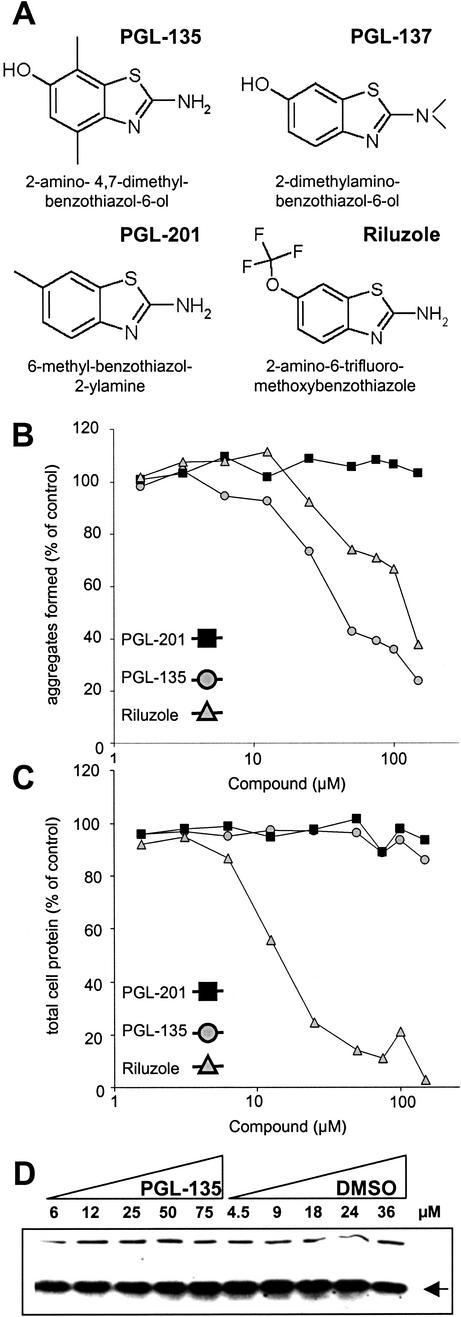
We tested more than 100 benzothiazole derivatives, including those identified by the high-throughput in vitro screen (Table 1), as well as several structural analogues obtained by computer analysis. Unexpectedly, we found that the most potent compounds in the cell-free assays—such as PGL-001, PGL-005, PGL-016, and PGL-034 (Figs. 2 and 3; Table 1)—were toxic for 293 Tet-Off cells (data not shown). Therefore, their activity on HDQ51 aggregation cannot be evaluated in this cell model system of HD. A similar result was also obtained, when the FDA-approved compound riluzole was tested in the cell-based assay (Fig. 4A). Increasing concentrations of riluzole markedly reduced the protein concentration, suggesting that the effect of this compound on HDQ51 aggregation is due to toxicity, but not due to an inhibition of this process (Fig. 4 B and C). In strong contrast, the benzothiazole derivatives PGL-135, PGL-137, and PGL-201 identified by computer analysis because of structural homology to the compounds shown in Table 1 were nontoxic for 293 Tet-Off cells (Fig. 4). We found that the compounds PGL-135 and PGL-137, but not PGL-201, inhibited HDQ51 aggregation in the cell-based assay. As shown in Fig. 4B, treatment of 293 Tet-Off cells with increasing concentrations of PGL-135 inhibited HDQ51 aggregation in a dose-dependent manner (IC50 value ≈40 μM). In comparison, PGL-137 suppressed HD exon 1 aggregation with an IC50 value of ≈100 μM (data not shown). Together, these results indicate that the benzothiazole derivatives PGL-135 and PGL-137 are nontoxic for 293 Tet-Off cells and inhibit HDQ51 aggregation in vivo.
Figure 4.
Inhibition of HD exon 1 aggregation in 293 Tet-Off cells. (A) Structure of chemical compounds counteracting HDQ51 aggregation in vivo. (B) Quantification of filter retardation assay results. Cells were incubated for 72 h in the presence of various concentrations of the indicated chemical compounds. Protein extracts were prepared and filtered through a cellulose acetate membrane; captured SDS-insoluble protein aggregates were detected with the HD1 antibody. The dots corresponding to the control reactions without added chemical compound (not shown) were arbitrarily set as 100. (C) Relative protein concentrations of the cell extracts analyzed in B. (D) Western blot of cell extracts prepared from 293 Tet-Off cells after treatment with increasing concentrations of the chemical compound PGL-135 and the solvent DMSO. The arrow indicates the HDQ51 monomer.
The effect of PGL-135 on the formation of HDQ51 aggregates in 293 Tet-Off cells was also examined by indirect immunofluorescence microscopy (Fig. 5A). Treatment of cells with PGL-135 significantly reduced the formation of large perinuclear inclusion bodies in a dose-dependent manner. Cultivation of the cells with 25 and 50 μM PGL-135 resulted in a 30 and 50% reduction of the average number of inclusion bodies per cell, respectively, consistent with the results obtained by the filter retardation assay (Fig. 4B).
Figure 5.
Immunofluorescence microscopy analysis. (A) 293 Tet-Off cells expressing Flag-tagged HD exon 1 protein with 51 glutamines (HDQ51) were cultivated for 72 h in the presence or absence of PGL-135. Formation of inclusion bodies with aggregated HDQ51 protein was followed by indirect immunofluorescence microscopy using the anti-Flag antibody. Inclusion bodies are indicated as red dots by the arrow. Nuclei were counterstained with Hoechst. A total of about 5,000 PGL-135-treated and untreated cells were examined. (B) Quantification of cells with inclusion bodies.
Discussion
In this study, we have developed cell-free and cell-based assays for the identification of chemical compounds that prevent the formation of SDS-insoluble HD exon 1 aggregates. Using a high-throughput in vitro filter retardation assay, we have identified about 300 chemical compounds in a library of ≈184,000 small molecules that suppress the accumulation of polyQ-containing huntingtin aggregates in a dose-dependent manner. Among these compounds, five larger groups with 5–35 structurally related small molecules were found. In this study, we have concentrated on the analysis of one group of small molecules containing 25 different benzothiazole derivatives.
Benzothiazoles are highly interesting molecules for drug development, because they already have been shown to be useful for treating cerebrovascular and neurodegenerative disorders. For example, riluzole extends the survival of neurons in amyotrophic lateral sclerosis patients (28, 29). It also has been tested for therapy of HD patients, where treatment with riluzole has positive effects on choreatic hyperkinesia (30). Moreover, it has been reported that riluzole leads to an extended lifespan in a mouse model of HD (31). Immunohistochemistry revealed profound changes in ubiquitination of HD-characteristic neuronal intranuclear inclusions, but no significant size reduction of huntingtin-positive inclusions was observed, indicating that this molecule does not directly interfere with huntingtin aggregation (31).
Our data show that benzothiazole derivatives are potent inhibitors of HD exon 1 aggregation in vitro and in cell culture model systems of HD. However, the mechanism of action of these molecules on a molecular level is unclear. We have shown previously that polyQ-containing HD exon 1 aggregates are formed by a nucleation-dependent polymerization (32). Thus, aggregation does not proceed immediately but after a lag phase during which monomers slowly assemble into unstable oligomeric structures (nuclei). Once these structures have formed, the addition of further monomers becomes thermodynamically favorable, resulting in the rapid accumulation of large fibrils (4). Currently, the structure of the huntingtin oligomers and fibrils is unknown; however, it is likely that both structures consist of cross β-sheets (4, 32).
We propose that benzothiazoles slow down HD exon 1 aggregation because they bind to polyQ-containing β-sheet structures. Similar to Congo red, they may interfere with nucleus formation and/or fibril growth. However, detailed in vitro drug-protein binding studies will be necessary to address these questions in more detail. Currently, it is not known whether benzothiazole derivatives such as PGL-001 preferentially interact with monomers, oligomers, or fibrillar structures. We have found that PGL-001 inhibits the aggregation of HDQ51 protein (conc. 0.625 μM) with an IC50 of 1.2 μM, suggesting that a protein-inhibitor stoichiometry of 1:2 is necessary and sufficient to inhibit HD exon 1 aggregation in vitro.
The activity of benzothiazoles to prevent huntingtin aggregation was also assessed in a cell culture model of HD. We found that, at conc. of 10–150 μM, the most potent compounds in the cell-free assays—such as PGL-001 or PGL-034—as well as the FDA-approved drug riluzole (Fig. 4), are toxic for 293 Tet-Off cells. Thus, the impact of these compounds on HD exon 1 aggregation in vivo cannot be determined. In comparison, the structurally related benzothiazole derivatives PGL-135 and PGL-137, identified by computer analysis, were nontoxic in the cell-based assay under the same conditions. We suggest that the compounds PGL-135 and PGL-137, similar to the most active compounds in the cell-free assay, directly interact with mutant HD exon 1 protein and thereby slow down the formation of insoluble protein aggregates in vivo. However, at this stage, we cannot exclude an indirect mode of action in the cell culture model of HD. For example, misfolding and aggregation of polyQ-containing HD exon 1 protein could be prevented by stimulation of a heat shock response or activation of the ubiquitin/proteasome system (11, 15, 16). Experiments to address these questions are needed.
Our studies show that the automated filter retardation assay described above is suitable for high-throughput screening of chemical compounds that prevent aggregate formation. Using this cell-free assay, very large compound libraries can be screened because the system is fast, robust, and relatively cheap. Furthermore, the amount needed of each substance for the aggregation reaction is small compared with other in vitro aggregation drug screening assays (34). Another strength of this assay is its ability to detect not only compounds that inhibit fibril growth (35), but also compounds that prevent nucleation.
A basic problem with drug screenings is that they tend to yield different results in cell-free, as opposed to cell-based, assays. Many compounds that show beneficial activities in the cell-free experiments are inactive or toxic in cell culture. We, e.g., found that benzothiazole derivatives that showed the greatest inhibitory effect in vitro were toxic in the cell assays. This finding means that the first step in screening can be to some extent misleading and that only the combination of cell-free and cell-based assays will result in the successful identification of small molecules that have the potential to be further developed into effective drugs. The challenge is to find chemical compounds that are nontoxic, that have a reasonable brain permeability, and that prevent the formation of huntingtin aggregates in the patient's neurons.
We have described a previously uncharacterized class of chemical compounds that prevent the formation of HD exon 1 aggregates in vitro and are biologically active in cell cultures. Now, the inhibitory activity of these molecules will be demonstrated in transgenic mouse models of HD and ultimately in humans.
Acknowledgments
We thank K. Genser, A. Dröge, S. Schnögl, and E. Scherzinger for critical reading of the manuscript. This work has been supported by grants from the Deutsche Forschungsgemeinschaft (WA1151/1-2 and WA1151/2-1), the Huntington's Disease Society of America, the Human Frontier Science Program Organization, and the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie (BioFuture Project 0311853).
Abbreviations
- HD
Huntington's disease
- polyQ
polyglutamine
- MALDI-TOF
matrix-assisted laser desorption ionization–time-of-flight
- GST-HD51
GSE-HD exon 1 fusion protein with 51 glutamines
- HDQ51
Flag-tagged HD exon 1 protein with 51 glutamines
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Self-Perpetuating Structural States in Biology, Disease, and Genetics,” held March 22–24, 2002, at the National Academy of Sciences in Washington, DC.
References
- 1.Harper P S. Huntington's Disease. London: Saunders; 1991. [Google Scholar]
- 2.Rubinsztein D C, Leggo J, Coles R, Almqvist E, Biancalana V, Cassiman J J, Chotai K, Connarty M, Crauford D, Curtis A, et al. Am J Hum Genet. 1996;59:16–22. [PMC free article] [PubMed] [Google Scholar]
- 3.Sathasivam K, Amaechi I, Mangiarini L, Bates G. Hum Genet. 1997;99:692–695. doi: 10.1007/s004390050432. [DOI] [PubMed] [Google Scholar]
- 4.Wanker E E. Biol Chem. 2000;381:937–942. doi: 10.1515/BC.2000.114. [DOI] [PubMed] [Google Scholar]
- 5.Paulson H L. Am J Hum Genet. 1999;64:339–345. doi: 10.1086/302269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wanker E E. Mol Med Today. 2000;6:387–391. doi: 10.1016/s1357-4310(00)01761-5. [DOI] [PubMed] [Google Scholar]
- 7.Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, Bates G P, Davies S W, Lehrach H, Wanker E E. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- 8.Krobitsch S, Lindquist S. Proc Natl Acad Sci USA. 2000;97:1589–1594. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faber P W, Alter J R, MacDonald M E, Hart A C. Proc Natl Acad Sci USA. 1999;96:179–184. doi: 10.1073/pnas.96.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies S W, Turmaine M, Cozens B A, DiFiglia M, Sharp A H, Ross C A, Scherzinger E, Wanker E E, Mangiarini L, Bates G P. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 11.Waelter S, Boeddrich A, Lurz R, Scherzinger E, Lueder G, Lehrach H, Wanker E E. Mol Biol Cell. 2001;12:1393–1407. doi: 10.1091/mbc.12.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamamoto A, Lucas J J, Hen R. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- 13.Lecerf J M, Shirley T L, Zhu Q, Kazantsev A, Amersdorfer P, Housman D E, Messer A, Huston J S. Proc Natl Acad Sci USA. 2001;98:4764–4769. doi: 10.1073/pnas.071058398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagai Y, Tucker T, Ren H Z, Kenan D J, Henderson B S, Keene J D, Strittmatter W J, Burke J R. J Biol Chem. 2000;275:10437–10442. doi: 10.1074/jbc.275.14.10437. [DOI] [PubMed] [Google Scholar]
- 15.Sittler A, Lurz R, Lueder G, Priller J, Hayer-Hartl M K, Hartl F U, Lehrach H, Wanker E E. Hum Mol Genet. 2001;10:1307–1315. doi: 10.1093/hmg/10.12.1307. [DOI] [PubMed] [Google Scholar]
- 16.Muchowski P J, Schaffar G, Sittler A, Wanker E E, Hayer-Hartl M K, Hartl F U. Proc Natl Acad Sci USA. 2000;97:7841–7846. doi: 10.1073/pnas.140202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heiser V, Scherzinger E, Boeddrich A, Nordhoff E, Lurz R, Schugardt N, Lehrach H, Wanker E E. Proc Natl Acad Sci USA. 2000;97:6739–6744. doi: 10.1073/pnas.110138997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bard F, Cannon C, Barbour R, Burke R L, Games D, Grajeda H, Guido T, Hu K, Huang J, Johnson-Wood K, et al. Nat Med. 2000;6:916–919. doi: 10.1038/78682. [DOI] [PubMed] [Google Scholar]
- 19.Soto C, Sigurdsson E M, Morelli L, Kumar R A, Castano E M, Frangione B. Nat Med. 1998;4:822–826. doi: 10.1038/nm0798-822. [DOI] [PubMed] [Google Scholar]
- 20.Korth C, May B C, Cohen F E, Prusiner S B. Proc Natl Acad Sci USA. 2001;98:9836–9841. doi: 10.1073/pnas.161274798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tatzelt J, Prusiner S B, Welch W J. EMBO J. 1996;15:6363–6373. [PMC free article] [PubMed] [Google Scholar]
- 22.Klunk W E, Debnath M L, Koros A M, Pettegrew J W. Life Sci. 1998;63:1807–1814. doi: 10.1016/s0024-3205(98)00454-8. [DOI] [PubMed] [Google Scholar]
- 23.Howlett D R, Perry A E, Godfrey F, Swatton J E, Jennings K H, Spitzfaden C, Wadsworth H, Wood S J, Markwell R E. Biochem J. 1999;340:283–289. [PMC free article] [PubMed] [Google Scholar]
- 24.Tomiyama T, Shoji A, Kataoka K, Suwa Y, Asano S, Kaneko H, Endo N. J Biol Chem. 1996;271:6839–6844. doi: 10.1074/jbc.271.12.6839. [DOI] [PubMed] [Google Scholar]
- 25.Wanker E E, Scherzinger E, Heiser V, Sittler A, Eickhoff H, Lehrach H. Methods Enzymol. 1999;309:375–386. doi: 10.1016/s0076-6879(99)09026-6. [DOI] [PubMed] [Google Scholar]
- 26.Sittler A, Walter S, Wedemeyer N, Hasenbank R, Scherzinger E, Eickhoff H, Bates G P, Lehrach H, Wanker E E. Mol Cell. 1998;2:427–436. doi: 10.1016/s1097-2765(00)80142-2. [DOI] [PubMed] [Google Scholar]
- 27.Jimonet P, Audiau F, Barreau M, Blanchard J C, Boireau A, Bour Y, Coleno M A, Doble A, Doerflinger G, Huu C D, et al. J Med Chem. 1999;42:2828–2843. doi: 10.1021/jm980202u. [DOI] [PubMed] [Google Scholar]
- 28.Bensimon G, Lacomblez L, Meininger V. N Engl J Med. 1994;330:585–591. doi: 10.1056/NEJM199403033300901. [DOI] [PubMed] [Google Scholar]
- 29.Lacomblez L, Bensimon G, Leigh P N, Guillet P, Meininger V. Lancet. 1996;347:1425–1431. doi: 10.1016/s0140-6736(96)91680-3. [DOI] [PubMed] [Google Scholar]
- 30.Rosas H D, Koroshetz W J, Jenkins B G, Chen Y I, Hayden D L, Beal M F, Cudkowicz M E. Movement Disorders. 1999;14:326–330. doi: 10.1002/1531-8257(199903)14:2<326::aid-mds1019>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 31.Schiefer J, Landwehrmeyer G B, Lüesse H-G, Sprünken A, Puls C, Milkereit A, Milkereit E, Kosinski C M. Movement Disorders. 2002;17:748–757. doi: 10.1002/mds.10229. [DOI] [PubMed] [Google Scholar]
- 32.Scherzinger E, Sittler A, Schweiger K, Heiser V, Lurz R, Hasenbank R, Bates G P, Lehrach H, Wanker E E. Proc Natl Acad Sci USA. 1999;96:4604–4609. doi: 10.1073/pnas.96.8.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gossen M, Bujard H. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin Y-M, Raffen R, Zhou Y, Cassidy C S, Flavin M T, Stevens F J. Amyloid. 2001;8:182–193. doi: 10.3109/13506120109007361. [DOI] [PubMed] [Google Scholar]
- 35.Esler W P, Stimson E R, Ghilardi J R, Felix A M, Lu Y-A, Vinters H V, Mantyh P W, Maggio J E. Nat Biotechnol. 1997;15:258–263. doi: 10.1038/nbt0397-258. [DOI] [PubMed] [Google Scholar]