Abstract
The synthesis of RNA–DNA primer by primosome requires coordination between primase and DNA polymerase α subunits, which is accompanied by unknown architectural rearrangements of multiple domains. Using cryogenic electron microscopy, we solved a 3.6 Å human primosome structure caught at an early stage of RNA primer elongation with deoxynucleotides. The structure confirms a long-standing role of primase large subunit and reveals new insights into how primosome is limited to synthesizing short RNA–DNA primers.
The human primosome, a complex of primase and DNA polymerase α (Polα), synthesizes the RNA–DNA primers that initiate DNA replication1,2. It is also involved in other DNA transactions, such as telomere maintenance3–5, DNA damage repair6 and innate immunity7–9. The biomedical importance of the primosome is evident with it being an emerging target for anticancer therapy10. The primosome holds a unique functional role in molecular biology due to its ability to de novo synthesize a short RNA primer by primase, composed of catalytic p49 and large p58 subunits, and then switching it to the two-subunit (catalytic p180 and accessory p70) Polα complex for extension with DNA1,2,11.
Structure–function studies of the primosome have provided much understanding on how the enzyme performs its catalytic actions, but most of these findings are limited to the context of individual domains or subunits, or subcomplexes12–21. Clearly, to understand how primosome makes the RNA–DNA primer, it is important to study the enzyme as a heterotetrametric complex. However, it is understandably challenging to elucidate structural details of the flexibly tethered primosome complex22,23. Indeed, till recently, only the apo state structure of the human primosome has been solved20,24. The enzyme apo state revealed an inhibitory compact structure that possibly explains how the primase domain can act first on a template1. A recent cryogenic electron microscopy (cryo-EM) structure of the human primosome caught in its preinitiation state with its single-stranded DNA-binding accessory protein, CST, provides critical insights into the enzyme’s RNA primer synthesis mechanism25. Despite these recent advances, the structural details of the primosome in its DNA elongation state remain unclear. In this Brief Communication, we report the cryo-EM structures of the human primosome trapped in the elongation state, which reveals an architecture different from the enzyme apo20 or preinitiation25 states and provides new insights into how the enzyme rearranges to accommodate RNA–DNA primer elongation and facilitate DNA synthesis termination.
Structures of human primosome DNA elongation complexes
We reconstituted the elongation complex by incubating a recombinant human primosome complex with dATP and a DNA template annealed to the 12-mer RNA–DNA primer that has a dideoxycytidine at the 3′-end to prevent further primer extension (Supplementary Table 1). Single-particle cryo-EM analysis yielded two major elongation complexes: a p180core–p58C binary complex (elongation complex I, EC-I; Fig. 1a) and an intact four-subunit complex (elongation complex II, EC-II; Fig. 1b), both bound to the template:primer. They were solved at 3.4 and 3.6 Å global resolution for EC-I and EC-II, respectively, allowing us to determine their atomic models (Fig. 1a,b, Table 1 and Extended Data Figs. 1 and 2).
Fig. 1|. Architecture of human primosome elongation complexes.
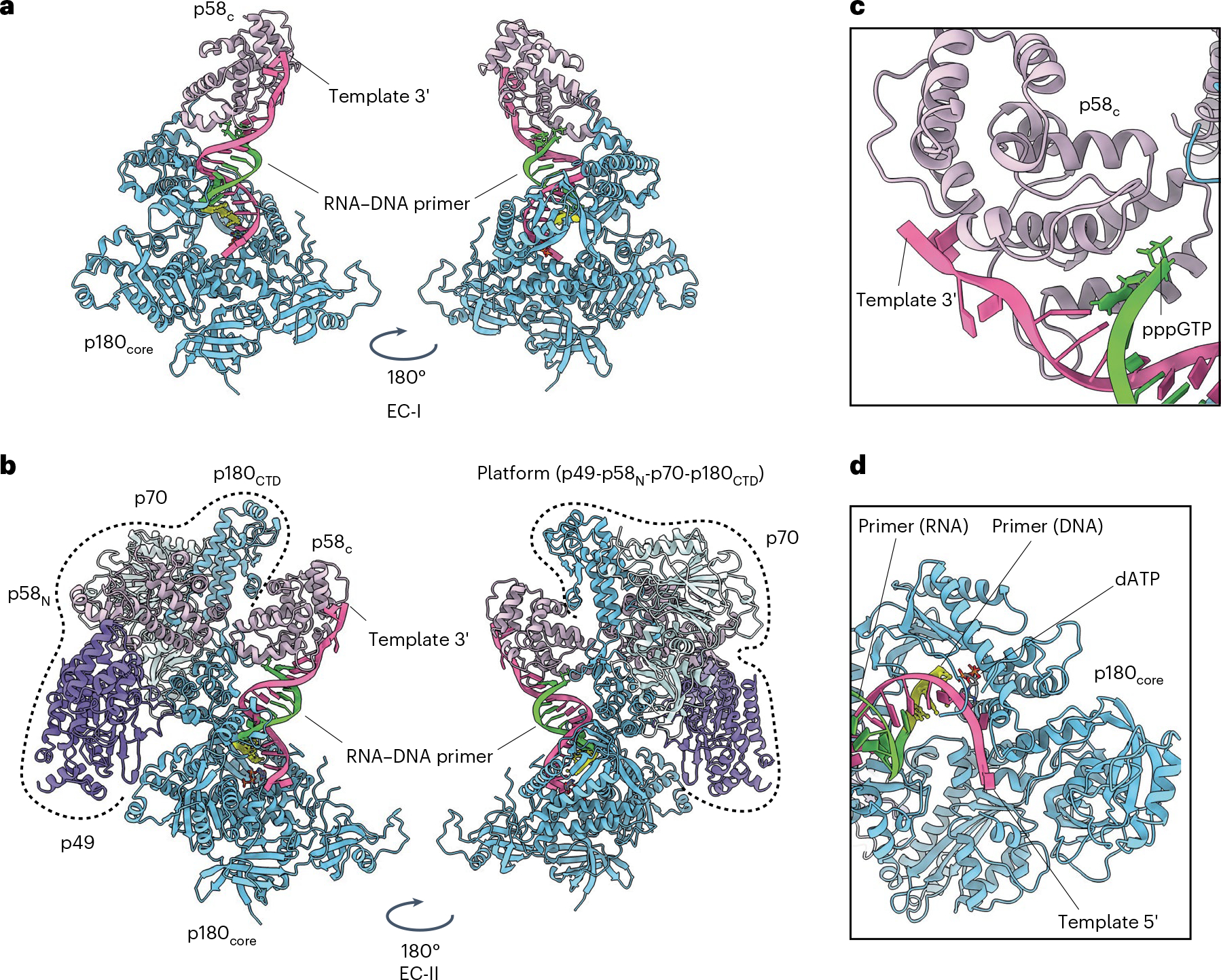
a, Atomic model of EC-I that consists of polymerase α catalytic domain (p180core) co-binding to the template:primer substrate with primase large subunit C-terminal domain (p58C). Template (5′-ATAATGGTCGTGCCGCCAATAA-3′) is colored as hot pink, while the RNA–DNA primer (5′-pppGGCGGCACGAC/ddC/−3′) is colored as lawn green (RNA) and yellow (DNA). Underlined sequence indicates primer complementary region on the template. RNA portion of the primer is italicized. ddC, dideoxycytidine. b, Atomic model of EC-II, consisting of all four primosome subunits: p49, p58, p70 and p180. The p180core and p58C domains bind the template:primer substrate in the same way as EC-I (see a). The platform domain (p49–p58N–p70–p180CTD) is encircled by dashed lines. c,d, Zoomed-in views of primer–template interactions by the p180core and p58C domains in the EC-II structure: the p58C domain engaged the 5′ triphosphate group of the RNA primer and the 3′ tail of the DNA template (c); the RNA–DNA primer reaches into the p180 catalytic center and is poised to react with incoming dATP (d).
Table 1|.
Cryo-EM data collection, refinement and validation statistics
| Primosome elongation complex-I (EMD-27256), (PDB8D96) | Primosome elongation complex-II (EMD-27258), (PDB8D9D) | |
|---|---|---|
| Data collection and processing | ||
| Magnification | 81,000x | 81,000x |
| Voltage (kV) | 300 | 300 |
| Electron exposure (e−Å−2) | 50.0 | 50.0 |
| Defocus range (μm) | −1.0 to −2.5 | −1.0 to −2.5 |
| Pixel size (Å) | 1.12 | 1.12 |
| Symmetry imposed | C1 | C1 |
| Initial particle images (no.) | 2,522,283 | 2,522,283 |
| Final particle images (no.) | 151,102 | 199,286 |
| Map resolution (Å) | 3.35 | 3.59 |
| FSC threshold | 0.143 | 0.143 |
| Map resolution range (Å) | 2.444 to 26.935 | 2.950 to 36.853 |
| Refinement | ||
| Initial model used (PDB code) | 5EXR, 4QCL | 5EXR, 4QCL |
| Model resolution (Å) | 3.4 (unmasked) | 3.6 (unmasked) |
| FSC threshold | 0.5 | 0.5 |
| Model resolution range (Å) | n/a | n/a |
| Map sharpening B factor (Å2) | −127.0 | −133.6 |
| Model composition | ||
| Non-hydrogen atoms | 9,204 | 19,442 |
| Protein residues | 1,056 | 2,312 |
| Ligands | 30 | 30 |
| B factors (Å2) | ||
| Protein | 154.19 | 94.70 |
| Ligand | 80.46 | 80.46 |
| r.m.s. deviations | ||
| Bond lengths (Å) | 0.002 | 0.003 |
| Bond angles (°) | 0.581 | 0.656 |
| Validation | ||
| MolProbity score | 1.53 | 1.68 |
| Clashscore | 7.36 | 7.57 |
| Poor rotamers (%) | 0 | 0 |
| Ramachandran plot | ||
| Favored(%) | 97.32 | 96.16 |
| Allowed (%) | 2.49 | 3.58 |
| Disallowed (%) | 0.19 | 0.26 |
N/a, not applicable; r.m.s, root-mean-square.
Both complexes bind the template:primer in similar fashion, with the p58C domain binding the primer’s 5′-triphosphate group while the p180core domain engages the primer 3′ end (Fig. 1c,d and Extended Data Fig. 3a). Comparison with the crystal structures of either template:primer-bound p180core (ref. 21) or template:primer-bound p58C (ref. 20) revealed that parts of the duplex are structurally well-conserved around areas interacting with the proteins, while some deviations are observed in remote areas (Extended Data Fig. 3b,c). Since EC-I and EC-II share the same template:primer interactions, it is possible that EC-I is a subcomplex of EC-II that was broken off during cryo-EM sample vitrification.
Our elongation complex structures provide the first direct evidence showing that the p58C domain remains bound to the template:primer after its handover from p49 to p180core. This continued primer 5′-end engagement by the p58C domain is consistent with past studies showing that this domain has high affinity to the template:primer (Kd ≈ 36 nM) (ref. 26), is essential for the internal primer handover27,28 and is compatible with a primer handover model that was generated without accessory proteins1,20. But then, why would the p58C domain persist in holding onto the template:primer after DNA elongation has started? We think this could be a fail-safe mechanism that prevents the catastrophic loss of the template:primer by primosome in an event when p180core releases the molecule.
Protein interactions underlying primer termination
The EC-II complex revealed that the platform domain, p49–p58N–p180C–p70, serves as a scaffold supporting the p180core–p58C–template:primer complex (Fig. 1a). Comparison with the apo state structure20 revealed a large rearrangement in the overall conformation of the primosome as it progressed to an elongation state (Extended Data Fig. 4). Within the platform domain, the disc-like p180C–p70 was rotated 37° relative to its position in the apo state (Extended Data Fig. 4b). The thumb subdomain of p180core is wedged between the p49 and p58N domains of primase, fixing the position of p180core relative to the platform (Fig. 2a).
Fig. 2|. Platform domain interaction with p180core is important for timely termination of RNA–DNA primer synthesis.
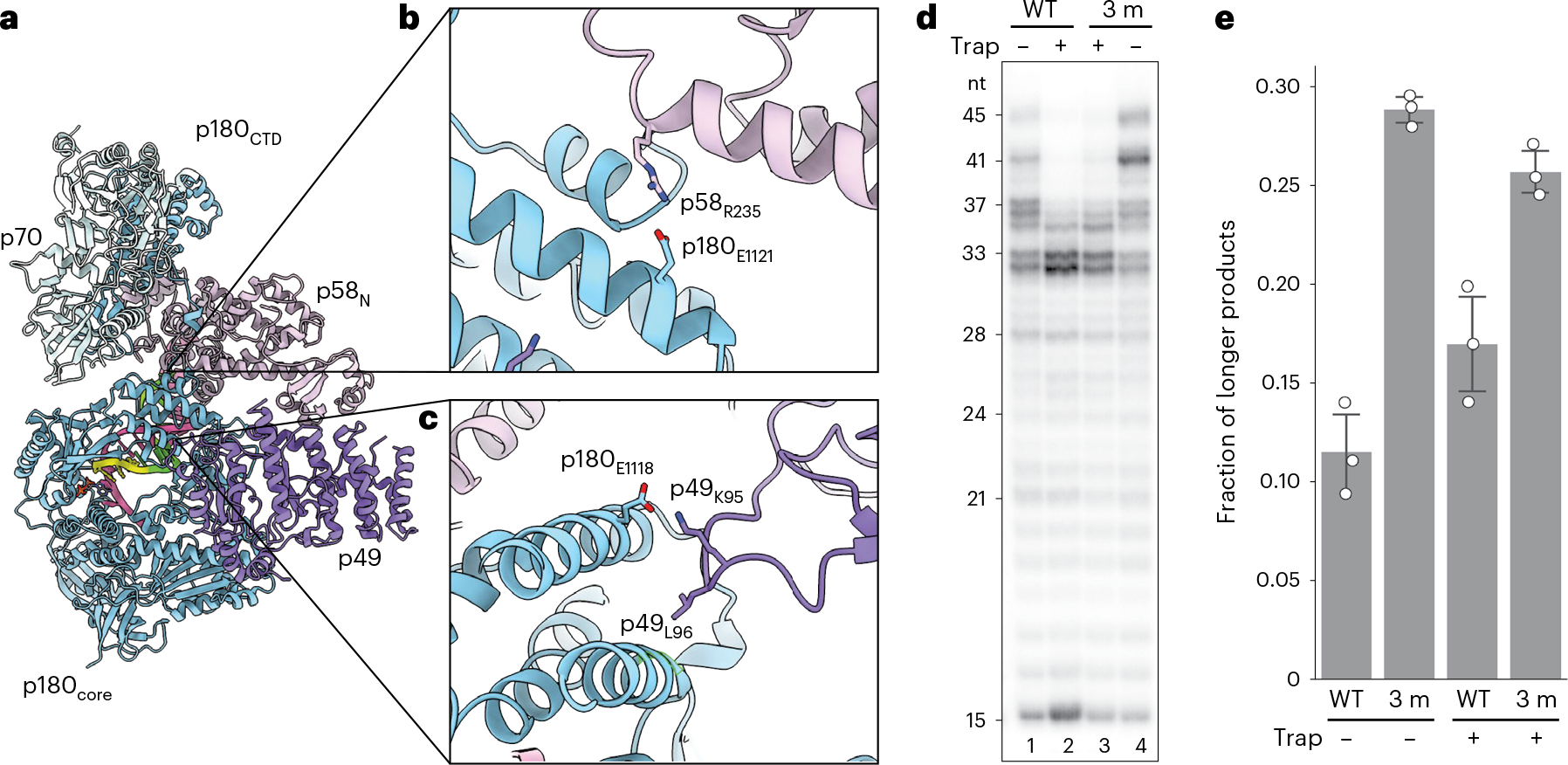
a, Intermolecular interactions enabling docking of p180core to the platform. Overall view is shown on the left. b,c, Close-up views of intersubunit interactions in the two interaction areas are shown on the right. The side chains of residues selected for mutations are shown and colored by heteroatom. d, Mutations disrupting the platform–p180core interaction increased processivity of DNA synthesis. The mutant primosome (ΔN-Polα/p49K95E/L96A/p58R235E) is termed 3 m. T1:P2 was used in all reactions. T1 and P2 sequences are provided in Supplementary Table 1. e, Processivity quantification of the results from d using fraction of longer products (defined as >37 nt products for lanes without trap and >33 nt products for lanes with trap added) over total products made. Independent data points (three independent experiments) are shown as hollow circles, while their mean ± standard deviation is drawn as columns and error bars, respectively.
Two new interaction sites, identified from the EC-II structure, seemingly support this novel elongation complex conformation (Fig. 2a–c). In one interaction area, interdomain hydrogen bonds are formed with participation of p49’s G97 backbone oxygen, and the side chains of K95 and Q100 with the side chains of S1127 and E1118 of p180core, respectively. In addition, the side chain of L96 from the p49 subunit is inserted into the hydrophobic pocket at the p180core surface (L1087, I1119, V1128 and hydrophobic parts of D1090 and T1091) (Fig. 2c). In another area, R235 of the p58N domain forms a salt bridge with E1121 of p180core; S113 of p58N interacts with D1242 and T1244 of p180core (Fig. 2b).
To explore the role of the newly identified platform–p180core interactions in primer DNA elongation, we introduced multiple point mutations in two subunits of primase, p49K95E/L96A and p58R235E, intended to disrupt p180core docking on the platform (Fig. 2d). Primer extension assay revealed that the mutations caused the enzyme to synthesize longer RNA–DNA primers, as compared with the wild-type (WT) version (Fig. 2d, compare lane 1 with lane 4, and quantified in Fig. 2e). This apparent increase in enzyme processivity was also observed at the single-turnover condition29 (Fig. 2d, compare lane 2 with lane 3, and quantified in Fig. 2e) and in reactions using primosomes containing full-length p180 (Extended Data Fig. 5).
Further isolation of the mutations (p49K95E/L96A at one interaction site and p58R235E at the other interaction site) yielded similar results (Extended Data Fig. 6), indicating that the residues K95 and L96 of p49 and R235 of p58 are indispensable for coordinating this new interaction between the platform and p180core. Interestingly, we found a similar increase in product length when either the p58C domain or the RNA’s triphosphate group is removed (Extended Data Fig. 7). These data imply that the template:primer–p58c interaction serves the same goal of limiting RNA–DNA primer length to ~35 nucleotides as the p180core–platform interactions that were identified in this work’s elongation complex structures.
Discussion
Our structure-guided mutation studies showed that the platform–p180core interaction of EC-II is important for preventing excessive DNA synthesis by Polα. But why would the enzyme evolve to throttle its primer elongation processivity? We believe this early termination behavior works in favor of the replication role of primosome, which is to seed multiple short primers for more processive DNA polymerases to take over. Without proof-reading capability30, excessive DNA elongation by primosome would lead to additional unnecessary mutations.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41594-023-00971-3.
Methods
Protein expression and purification
Expression and purification of the human primase heterodimer31 and primosome (p49–p58–p180–p70) (ref. 32) have been described elsewhere. In short, human primase was overexpressed in Escherichia coli strain Rosetta-2 (DE3) and purified in two steps using Heparin HP HiTrap (Cytiva) and monoQ (Cytiva) columns. Human Polα with a His-tag at the N-terminus of a second subunit was expressed in insect cells and purified in three steps using Ni-IDA (Bio-Rad), Heparin HP HiTrap (Cytiva) and hydroxyapatite (Bio-Rad) columns. The obtained primase and Polα samples were mixed at a molar ratio of 1.2:1, and the reconstituted primosome was purified by size-exclusion chromatography using the Superose 12 10/300 GL (Cytiva) column. In some primosome constructs, including the one used for structural studies, the N-terminus of p180 (residues 1–334) has been deleted because it is poorly folded33 and is not required for activity20 and interaction with other primosome subunits25. Mutations to p49 and p58 were made by standard site-directed mutagenesis34. The expression and purification of the full-length human primosome and mutants were performed according to an established protocol25. In brief, healthy Trichoplusia ni insect cells (Expression Systems) were infected with four baculoviruses (p180, p70, p58 and p49) for co-expression of the full-length primosome and its mutants. After 66–68 h, the infected T. ni cells were collected for protein purification. The primosome complexes were obtained using tandem affinity purification: Ni-NTA agarose resin (Qiagen) was used to pull down His-tagged p49, p58 and p70 proteins, and then the eluent was subjected to a second pull-down using Strep-Tactin XT 4Flow-resin (IBA LifeScience) for Strep-tagged p180. All protein complexes were verified using SDS–PAGE analysis.
Oligonucleotides for structural and functional studies
Sequences of all oligonucleotides are provided in Supplementary Table 1. Oligonucleotides without 5′-triphosphate were obtained from Integrated DNA Technologies. The 5-mer RNA primer P5 containing the 5′-triphosphate (5′-pppGGCGG) was obtained as described previously using DNA duplex T3:P4 and RNA polymerase of bacteriophage T7 (ref. 26). Template:primers containing P2 or P3 were obtained by ligation using the corresponding template, P5, RNA ligase 2 of bacteriophage T4 (New England BioLabs), and P6 or P7, respectively. Reactions were incubated for 1 h at 25 °C, and ligated duplexes were purified by 1 ml monoQ column (Cytiva) at 50 °C.
Sample preparation for cryo-EM studies
The primosome elongation complex was obtained by mixing recombinant human primosome with T2:P3 duplex at a molar ratio of 1:1 in a buffer containing 20 mM Tris–HCl, pH 7.7, 100 mM KCl, 1% (v/v) glycerol, 1 mM TCEP, 1.5 mM CaCl2 and 1.7 mM dATP. The reconstituted elongation complex was concentrated to 8 mg ml−1 and flash-frozen as 5 μl aliquots before storage in a −80 °C freezer. The frozen samples were thawed just before cryo-EM grid preparation. To prevent sample damage during cryo-EM grid vitrification, a final 8 mM CHAPSO35 was added to the elongation complex sample before applying 3.5 μl of the sample to a glow-discharged holey electron microscopy grid (Quantifoil QF 1.2/1.3, 300 mesh). The grid was blotted for 3.5 s at 4 °C and 95% relative humidity and plunge-frozen with liquid ethane as cryogen using the Vitrobot machine (FEI, ThermoFisher).
Cryo-EM data collection and processing
A total of 13,243 movies were collected on a Titan Krios transmission electron microscope (FEI, ThermoFisher) at the National Cryo-Electron Microscope Facility at the National Cancer Institute of the National Institutes of Health. The movies were imaged at a nominal pixel resolution of 1.12 Å per pixel using a K3 direct electron detector in counting mode and energy filter at 20 eV (Gatan). Each movie is collected across 40 frames with a total electron dose of 50 e− Å−2. The movies were aligned and their constant transfer function (CTF) parameters calculated using CryoSparc2 (ref. 36) patch motion correction and CTF estimation. A total of 12,168 micrographs remained after curation, and from these micrographs, 2,522,283 particles were picked and extracted at 4× binning (4.48 Å per pixel). The particles were subjected to a single round of 2D classification to remove most junk particles/ice contaminations. Selected particles (899,717 particles) were further sorted into four separate reference-free 3D classes using CryoSparc2 ab initio modeling. Two distinct conformations of the elongation complexes (EC-I and EC-II) were resolved in this step.
The particles from these two classes (total of 589,593 particles) were re-extracted at the original pixel size before undergoing another round of four-class ab initio modeling. This step yielded the final particles subsets for EC-I and EC-II. In total, 32.3% of the particles (151,102 particles) belong to EC-I while 41.9% of the particles (199,286 particles) belong to EC-II. Non-uniform refinement with per-particle CTF refinement37 resulted in a global resolution (reported at Fourier shell correlation of 0.143) of 3.4 Å for EC-I and 3.6 Å for EC-II.
Model building, refinement and validation
The EC-I and EC-II models were built by docking solved structures of individual domains20,21 into the cryo-EM maps. Docking was done using ChimeraX software38. The elongation complex models were inspected visually with respect to their fit in their respective cryo-EM maps and manually corrected using Coot39. The models were then subjected to real-space refinement using the Phenix software40. Cryo-EM map to model validation statistics was derived from Phenix’s comprehensive validation package.
Primer extension assay
Activity was tested in 10 μl reaction containing 0.6 μM template:primer, 0.2 μM enzyme, 10 μM dNTPs, 0.1 μM [α−32P]-dCTP (3,000 Ci mmol−1; PerkinElmer), 10 μM trap and buffer consisting of 30 mM Tris–HEPES, pH 7.8, 120 mM KCl, 30 mM NaCl, 1% glycerol, 2 mM TCEP, 5 mM MgCl2 and 0.2 mg ml−1 BSA. The trap was a T4:P8 duplex containing the dideoxy-cytosine at the primer 3′-end to make a dead-end complex with Polα. The enzyme was preincubated with a template:primer in 5 μl for 1 min on ice and for 10 s at 35 °C, then reaction was initiated by addition of 5 μl solution containing dNTPs and trap. Reactions were incubated in PCR tubes on a water bath for 30 s at 35 °C and stopped by mixing with equal volume of formamide loading buffer (90% v/v formamide, 50 mM EDTA, pH 8, and 0.02% bromophenol blue), heated at 95 °C for 1 min and resolved by 20% urea–PAGE (UreaGel System (19:1 acrylamide/bisacrylamide), National Diagnostics) for 2.5 h at 3,000 V. The reaction products were visualized by a phosphorimager (Typhoon FLA 9500, Cytiva). All activity gels were repeated at least three times for reproducibility. The activity analysis was carried out using the GelAnalyzer software (GelAnalyzer 19.1; www.gelanalyzer.com).
Extended Data
Extended Data Fig. 1|. Cryo-EM processing pipeline for the elongation complexes.
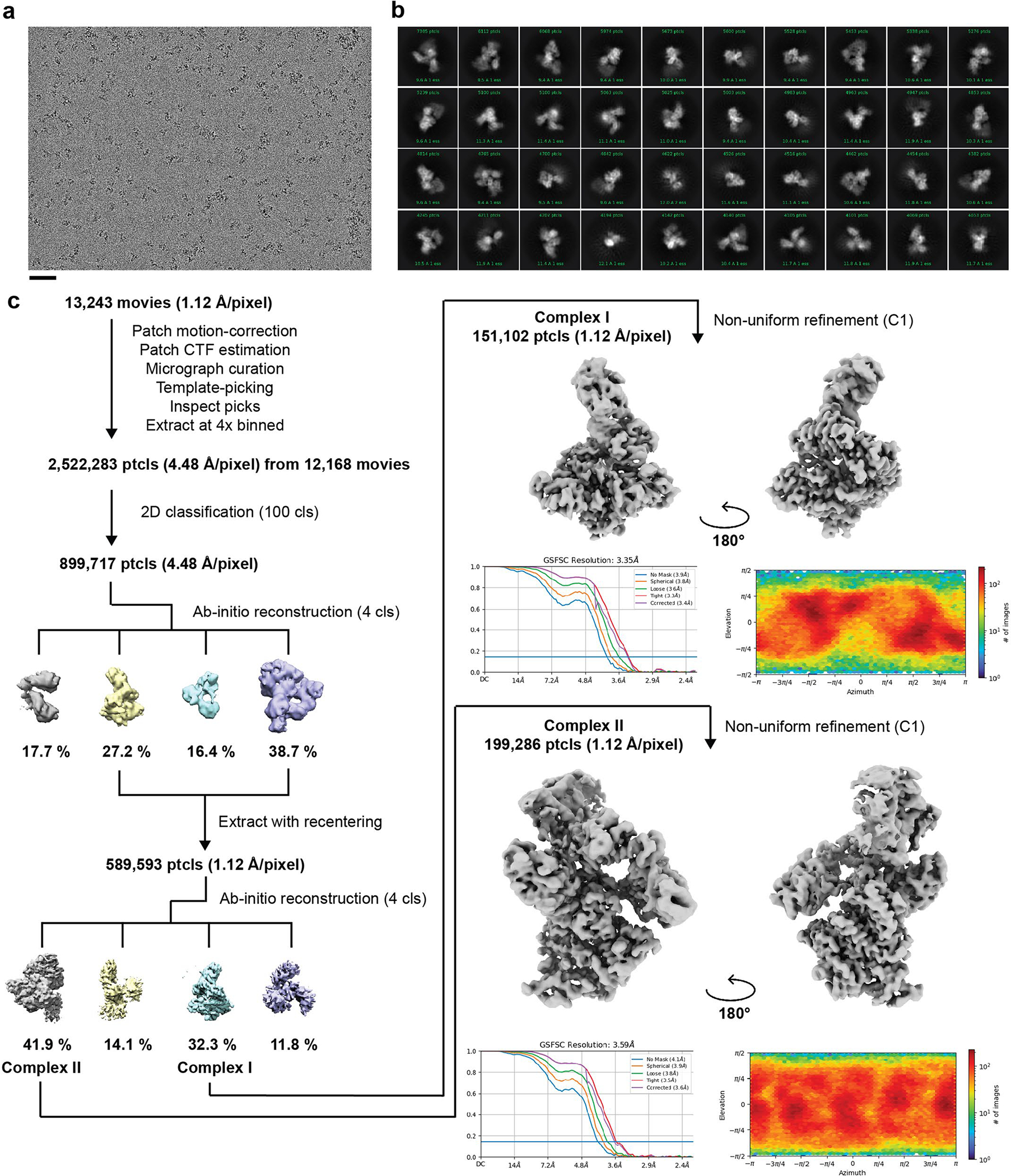
(a) Representative micrograph (n = 13,243) of the cryo-EM dataset. The scale bar dimension is 500 Å. (b) 2D classification averages of the elongation complexes. (c) Cryo-EM processing pipeline that was used to obtain the two cryo-EM maps of elongation complexes I and II.
Extended Data Fig. 2|. Local resolution maps and map to model comparison of the cryo-EM reconstruction.
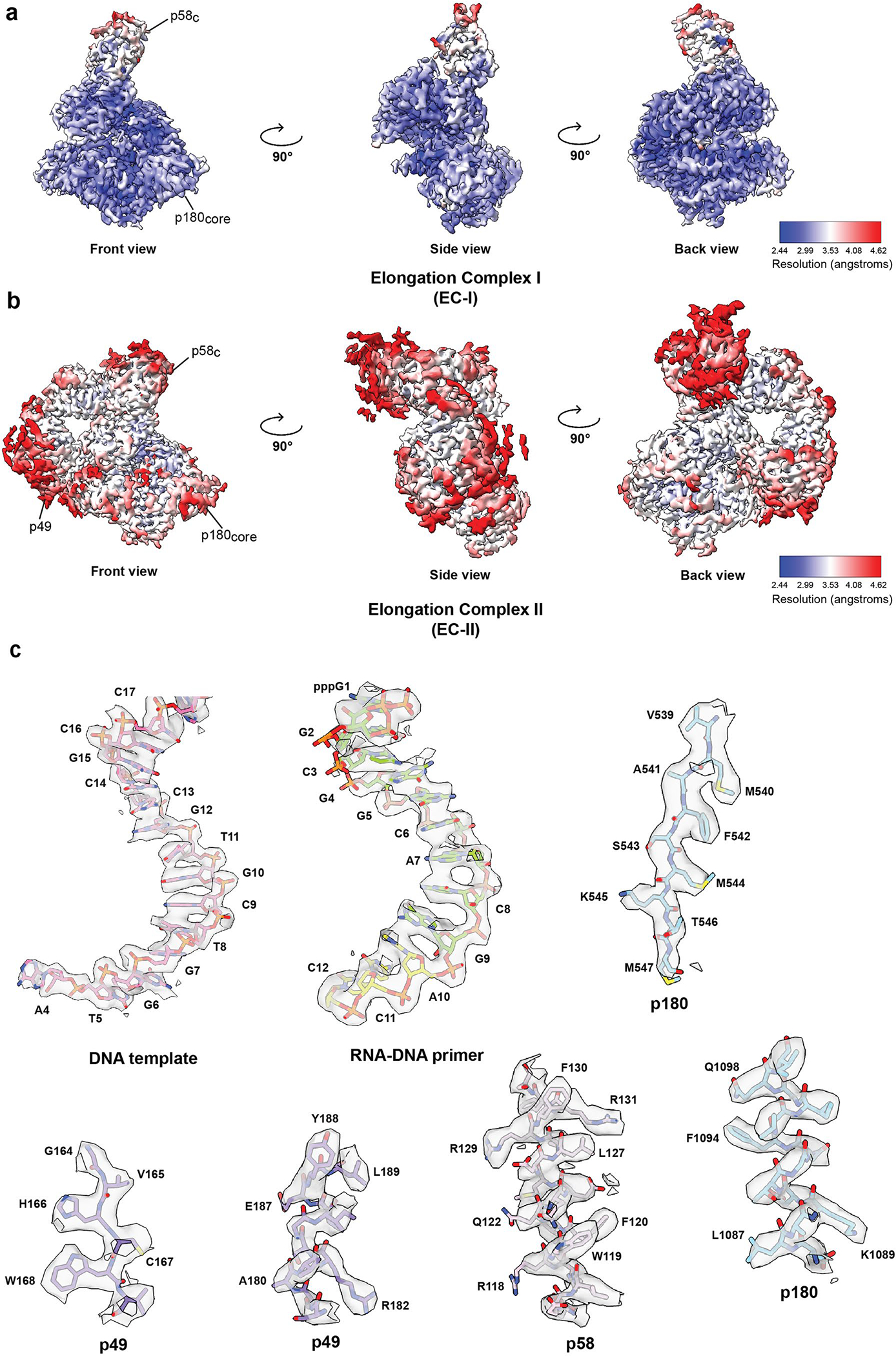
Local resolution maps of the (a) EC-I and (b) EC-II cryo-EM structures. (c) Representative cryo-EM densities encasing the corresponding atomic models of DNA template, RNA-DNA primer, p49, p58 and p180.
Extended Data Fig. 3|. Structural comparison of template:primer bound to Polα catalytic core and to p58C between the two cryo-EM elongation complex models.
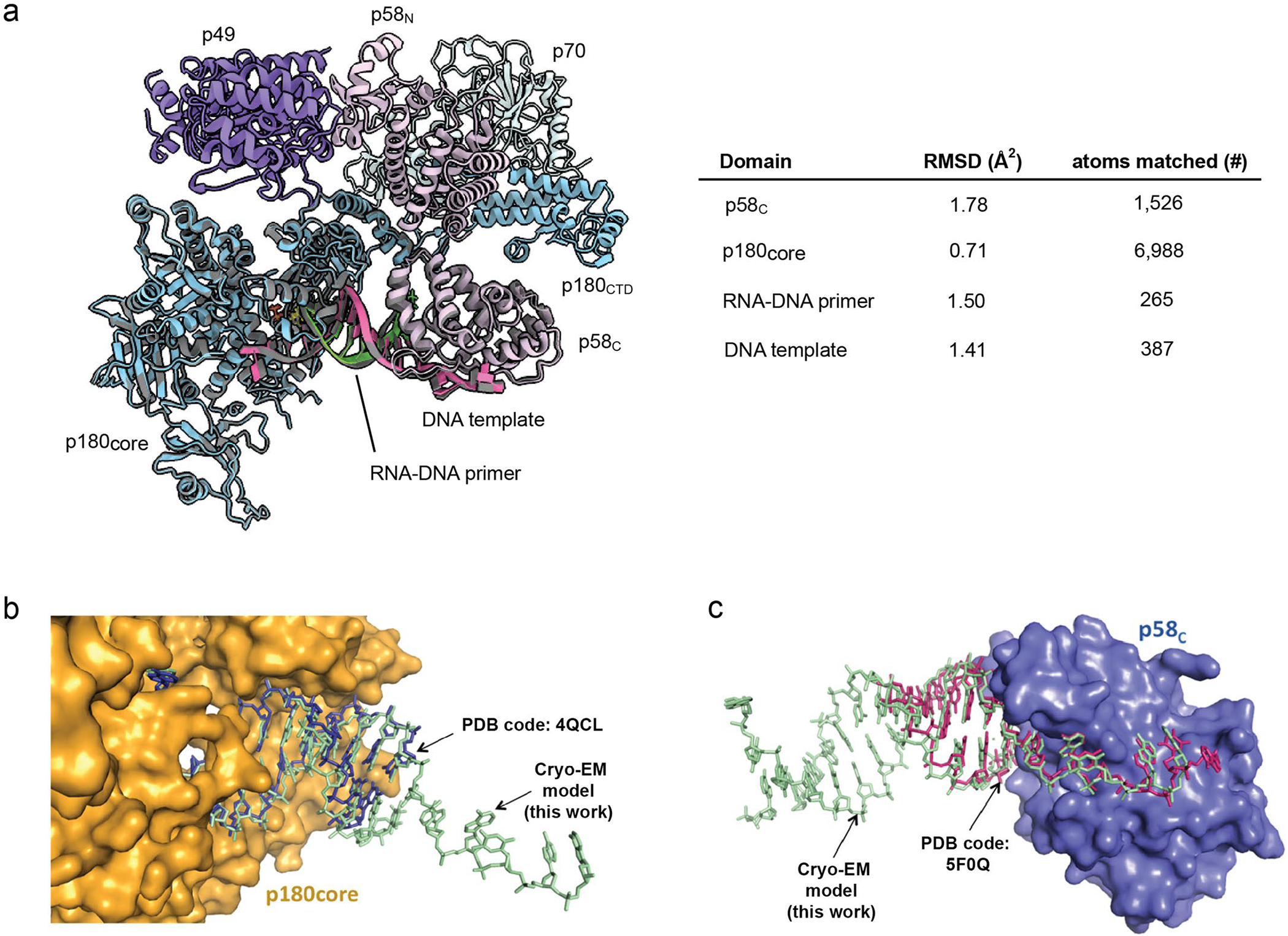
(a) The models of elongation complex I and II are aligned and the RMSD values of p58C, p180core, RNA-DNA primer, and DNA template are calculated. Their RMSD values are shown in the table in the panel. A ribbon model of the EC-II is shown with its subunits colored as per described in main text. The superimposed EC-I ribbon model is colored grey to illustrate the similarity between the two models. The ChimeraX software37 was used to perform the above RMSD analysis. (b) The crystal structure of p180core/template:primer (PDB code: 4QCL) was superimposed with p180core/template:primer part of primosome elongation complex II (RMSD = 1.10 Å2). (c) The crystal structure of template:primer/p58C (PDB code: 5F0Q) was superimposed with template:primer/p58C part of primosome elongation complex II (RMSD = 0.59 Å2).
Extended Data Fig. 4|. Primosome undergoes a large conformation change as it progresses from the apo to elongation state.
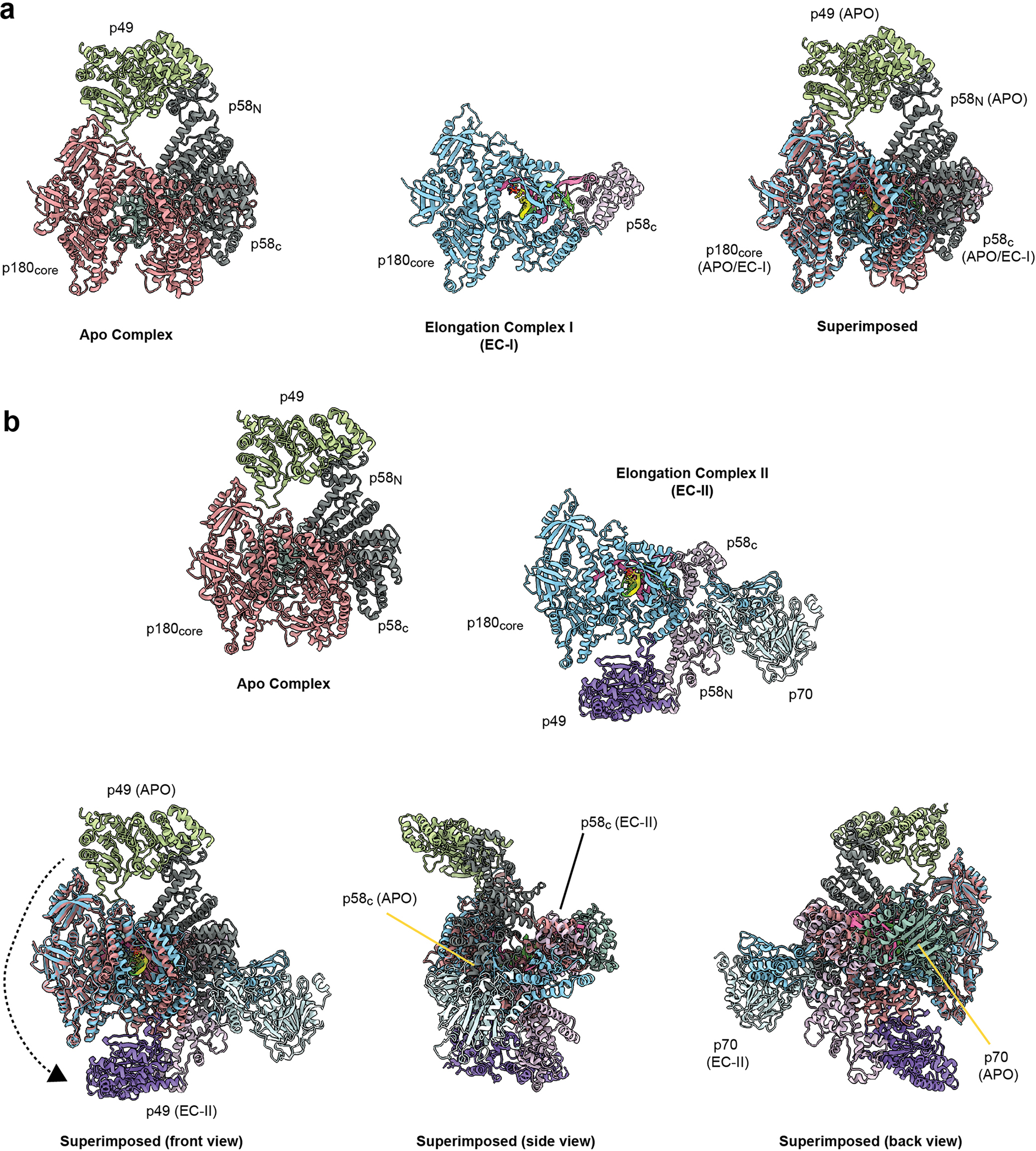
The apo state of human primosome (PDB code: 5EXR) is compared with this work’s elongation states primosome, (a) EC-I and (b) EC-II. The four subunits of APO primosome are colored: p49 as green, p58 as dark grey, p70 as olive green, and p180 as salmon red. The elongation state primosomes are colored as described in Fig. 1. The APO and EC-I/II structures are aligned using the p180 subunit. Visual inspection of APO vs EC-II state shows the p49, p58, and p70 domains are rearranged relative to the p180core domain.
Extended Data Fig. 5|. Mutagenesis analysis of platform-p180core interaction by primer extension assay using primosome with full-length Polα.
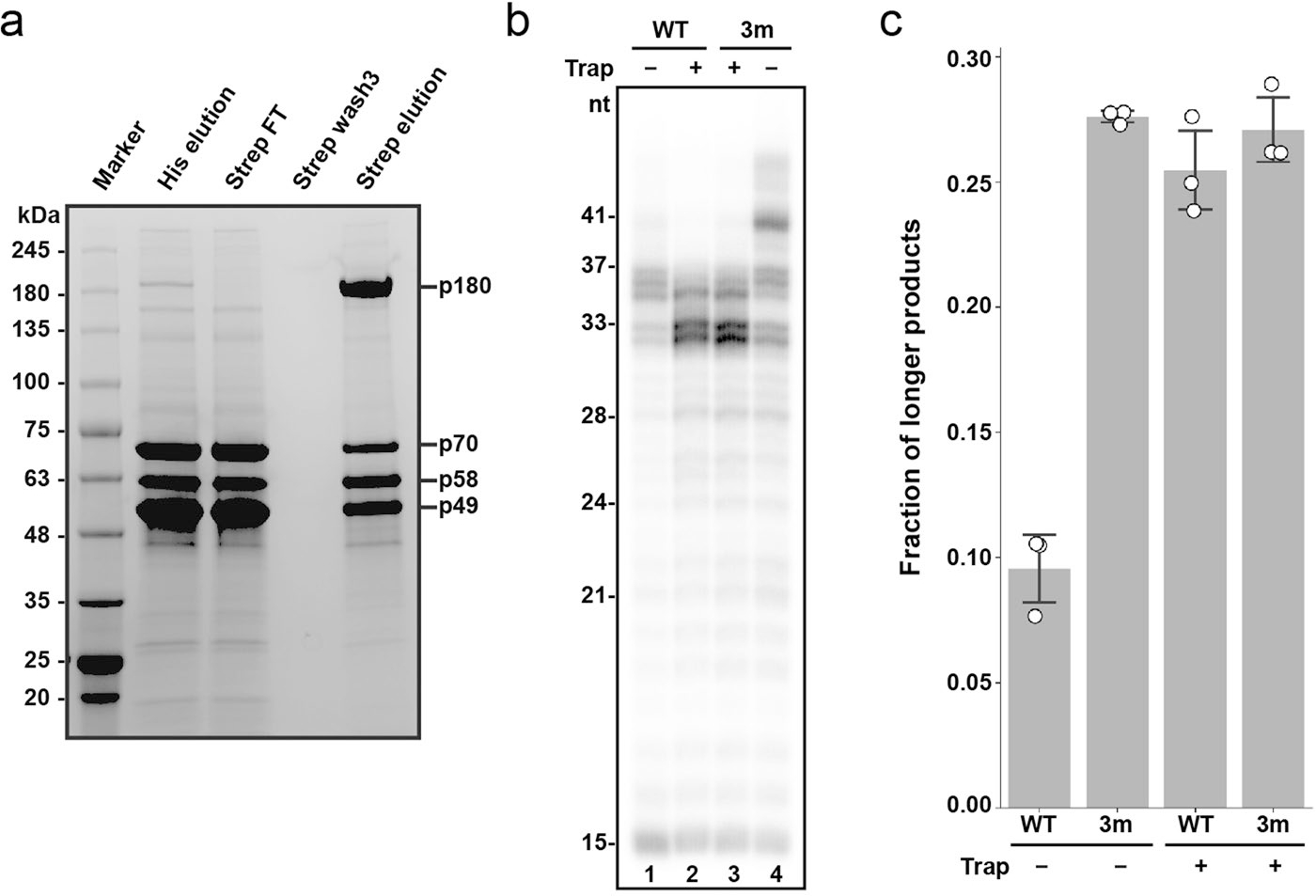
(a) SDS-PAGE analysis of tandem affinity purified full-length Primosome. 6xHis-tagged subunits (p49, p58, p70) were first enriched before pulling down the Twin-Strep-tagged p180 to obtain the assembled primosome complex. FT: flowthrough. The results are reproduced across multiple independent experiments (n > 3). (b) Mutations disrupting the platform-p180core interaction increase processivity of DNA synthesis. The mutant primosome (Polα/p49K95E/L96A/p58R235E), with full-length p180, is referred as 3 m in the panels. T1:P2 was used in all reactions. (c) Processivity quantification of the results from panel a using fraction of longer products (defined as >37nt products for -trap and >33nt products for +trap) over total product made. Triplicate data are represented as hollow circles while their mean and standard deviation (mean ± SD) are show as columns and error bars, respectively.
Extended Data Fig. 6|. Detailed mutagenesis analysis of the platform-p180core interaction.
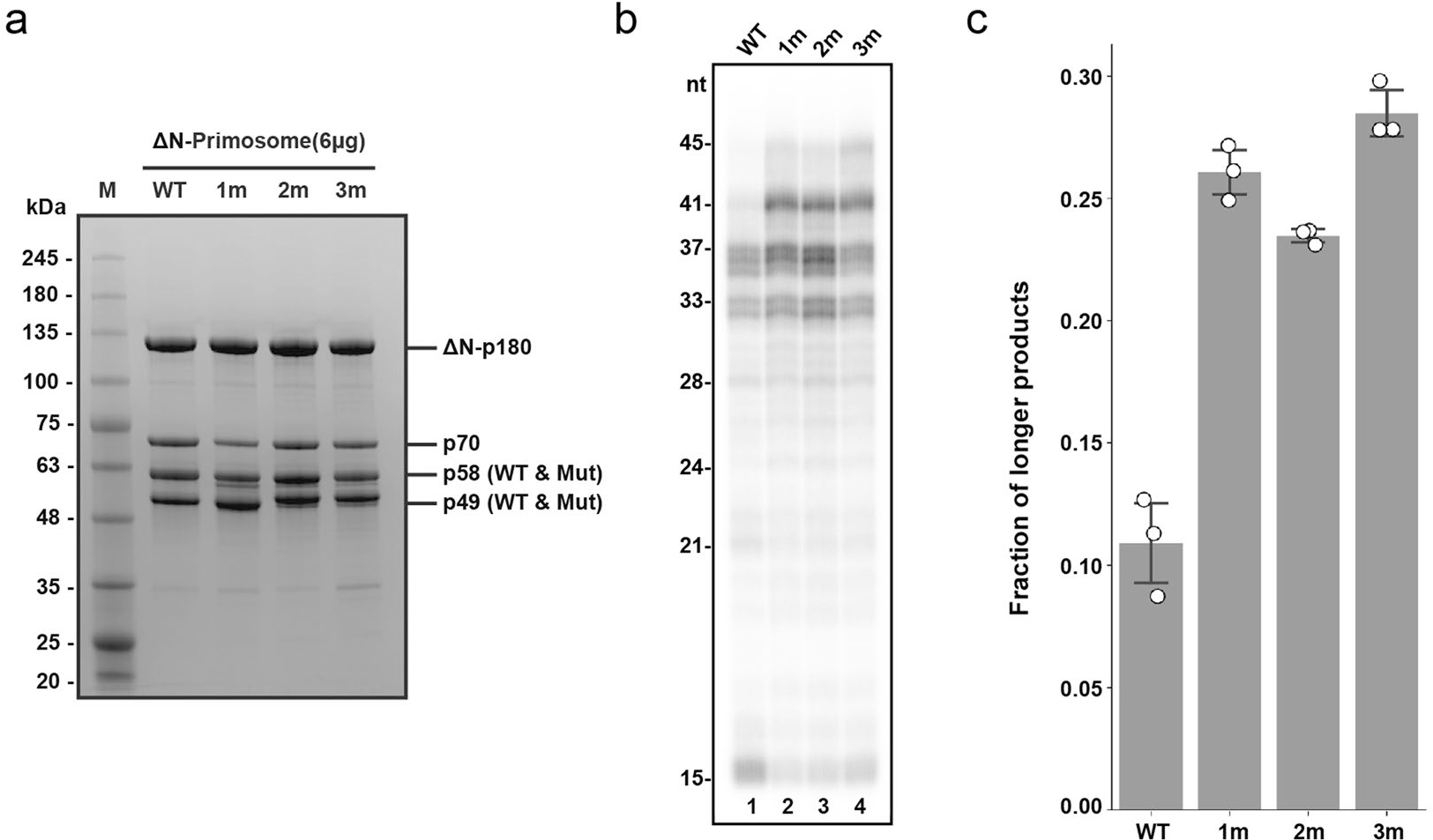
(a) Purified recombinant wild-type and mutant human primosomes. Mutant primosomes are annotated in gel as 1 m (ΔN-Polα/p49/p58R235E), 2 m (ΔN-Polα/p58/p49K95E/L96A), and 3 m (ΔN-Polα/p49K95E/L96A/p58R235E). This experiment was performed once. (b) Mutation(s) on either p49 or p58 has a similar effect as the combined mutation (3 m) on enzyme processivity. T1:P2 was used in all reactions without trap added. (c) Processivity quantification using fraction of longer products (>37nt) over total product from triplicates of panel b results. The triplicate data are represented as hollow circles while their mean and standard deviation (mean ± SD) are shown as columns and error bars.
Extended Data Fig. 7|. DNA synthesis by primosome upon disruption of the interaction between p58C and template:primer.
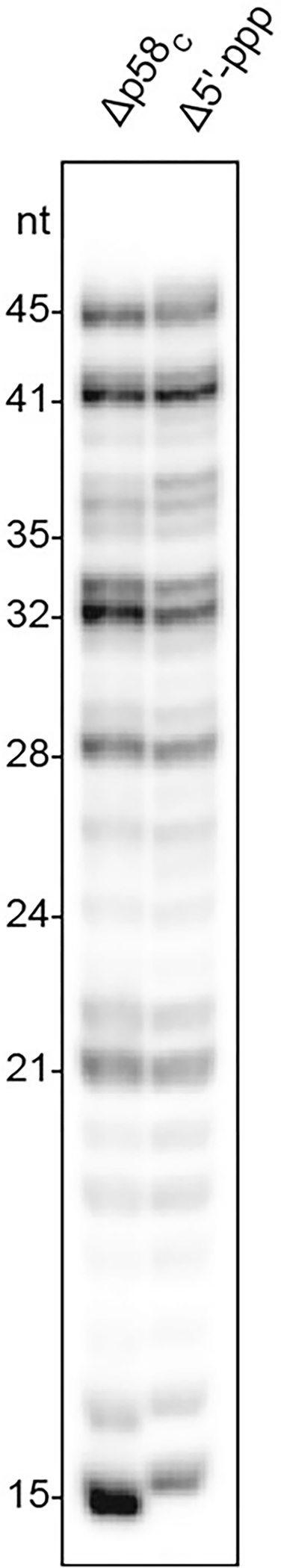
As compared to the RNA-DNA primers made by wild-type primosome using template:primer with 5’-triphosphate (see Fig. 2d, lane 1), reactions using primosome with deleted p58C or the template:primer without the 5’-triphosphate resulted in longer products being made by the enzyme. Reactions corresponding to the left and right lanes contain T1:P2 and T1-P1, respectively. The results are reproducible across multiple independent experiments (n = 3).
Supplementary Material
Acknowledgements
We thank members of the Lim lab and Tahirov lab for their helpful discussions. We also thank M. Cox and I. Rayment for their constructive feedback. We also thank Z. Xu for her assistance in figures preparation. This research was, in part, supported by the Cryo-EM Research Center in the Department of Biochemistry at the University of Wisconsin-Madison and the National Cancer Institute’s National Cryo-EM Facility at the Frederick National Laboratory for Cancer Research under contract HSSN261200800001E. This work was supported by the National Institute of General Medical Sciences grants R35GM127085 to T.H.T. and R00GM131023 to C.L. Support for this work was also provided to C.L. by the University of Wisconsin–Madison, Office of the Vice-Chancellor for Research and Graduate Education with funding from the Wisconsin Alumni Research Foundation and the Department of Biochemistry.
Footnotes
Competing interests
The authors declare no competing interests.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s41594-023-00971-3.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41594-023-00971-3.
Peer review information Nature Structural & Molecular Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Sara Osman was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team. Peer reviewer reports are available.
Reprints and permissions information is available at www.nature.com/reprints.
Data availability
The cryo-EM maps of the human primosome elongation complexes have been deposited to the Electron Microscopy Data Bank (EMDB) under the accession code EMD-27256 (EC-I) and EMD-27258 (EC-II). The corresponding atomic coordinates are deposited in the RSCB Protein Data Bank under the accession code 8D96 (EC-I) and 8D9D (EC-II). The coordinates for the initial models used are available in the PDB under accession codes 5EXR (human p49, p58 and p70) and 4QCL (p180 and template:primer). Source data are provided with this paper.
References
- 1.Baranovskiy AG & Tahirov TH Elaborated action of the human primosome. Genes 8, 62 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pellegrini L The Pol alpha-primase complex. Subcell. Biochem 62, 157–169 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Lim C & Cech TR Shaping human telomeres: from shelterin and CST complexes to telomeric chromatin organization. Nat. Rev. Mol. Cell Biol. 22, 283–298 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan X & Price CM Coordinate regulation of G- and C strand length during new telomere synthesis. Mol. Biol. Cell 8, 2145–2155 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reveal PM, Henkels KM & Turchi JJ Synthesis of the mammalian telomere lagging strand in vitro. J. Biol. Chem. 272, 11678–11681 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Mirman Z et al. 53BP1-RIF1-shieldin counteracts DSB resection through CST- and Polα-dependent fill-in. Nature 560, 112–116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Starokadomskyy P et al. DNA polymerase-α regulates the activation of type I interferons through cytosolic RNA:DNA synthesis. Nat. Immunol. 17, 495–504 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kilkenny ML et al. Structural basis for the interaction of SARS-CoV-2 virulence factor nsp1 with DNA polymerase α-primase. Protein Sci. 31, 333–344 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang L, Sheraz M, McGrane M, Chang J & Guo JT DNA polymerase α is essential for intracellular amplification of hepatitis B virus covalently closed circular DNA. PLoS Pathog. 15, e1007742 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han T et al. The antitumor toxin CD437 is a direct inhibitor of DNA polymerase alpha. Nat. Chem. Biol. 12, 511–515 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doublie S & Zahn KE Structural insights into eukaryotic DNA replication. Front. Microbiol. 5, 444 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sauguet L, Klinge S, Perera RL, Maman JD & Pellegrini L Shared active site architecture between the large subunit of eukaryotic primase and DNA photolyase. PLoS ONE 5, e10083 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaithiyalingam S, Warren EM, Eichman BF & Chazin WJ Insights into eukaryotic DNA priming from the structure and functional interactions of the 4Fe-4S cluster domain of human DNA primase. Proc. Natl Acad. Sci. USA 107, 13684–13689 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agarkar VB, Babayeva ND, Pavlov YI & Tahirov TH Crystal structure of the C-terminal domain of human DNA primase large subunit: implications for the mechanism of the primase-polymerase alpha switch. Cell Cycle 10, 926–931 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kilkenny ML, Longo MA, Perera RL & Pellegrini L Structures of human primase reveal design of nucleotide elongation site and mode of Pol alpha tethering. Proc. Natl Acad. Sci. USA 110, 15961–15966 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perera RL et al. Mechanism for priming DNA synthesis by yeast DNA polymerase alpha. eLife 2, e00482 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baranovskiy AG et al. Structural basis for inhibition of DNA replication by aphidicolin. Nucleic Acids Res. 42, 14013–14021 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaithiyalingam S et al. Insights into eukaryotic primer synthesis from structures of the p48 subunit of human DNA primase. J. Mol. Biol. 426, 558–569 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baranovskiy AG et al. Crystal structure of the human primase. J. Biol. Chem. 290, 5635–5646 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baranovskiy AG et al. Mechanism of concerted RNA–DNA primer synthesis by the human primosome. J. Biol. Chem. 291, 10006–10020 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baranovskiy AG et al. Activity and fidelity of human DNA polymerase alpha depend on primer structure. J. Biol. Chem. 293, 6824–6843 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klinge S, Nunez-Ramirez R, Llorca O & Pellegrini L 3D architecture of DNA Pol alpha reveals the functional core of multi-subunit replicative polymerases. EMBO J. 28, 1978–1987 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nunez-Ramirez R et al. Flexible tethering of primase and DNA Pol α in the eukaryotic primosome. Nucleic Acids Res. 39, 8187–8199 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kilkenny ML et al. Structural basis for the interaction of SARS-CoV-2 virulence factor nsp1 with DNA polymerase α-primase. Protein Sci. 31, 333–344 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He Q et al. Structures of the human CST–Polα–primase complex bound to telomere templates. Nature 608, 826–832 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baranovskiy AG et al. Insight into the human DNA primase interaction with template–primer. J. Biol. Chem. 291, 4793–4802 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheaff RJ, Kuchta RD & Ilsley D Calf thymus DNA polymerase α-primase: ‘communication’ and primer–template movement between the two active sites. Biochemistry 33, 2247–2254 (1994). [DOI] [PubMed] [Google Scholar]
- 28.Arezi B, Kirk BW, Copeland WC & Kuchta RD Interactions of DNA with human DNA primase monitored with photoactivatable cross-linking agents: implications for the role of the p58 subunit. Biochemistry 38, 12899–12907 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Baranovskiy AG, Lisova AE, Morstadt LM, Babayeva ND & Tahirov TH Insight into RNA–DNA primer length counting by human primosome. Nucleic Acids Res. 50, 6264–6270 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reijns MAM et al. Lagging-strand replication shapes the mutational landscape of the genome. Nature 518, 502–506 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baranovskiy AG et al. Crystallization and preliminary X-ray diffraction analysis of human DNA primase. Acta Crystallogr. F 70, 206–210 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Baranovskiy AG, Tahirov TH & Pavlov YI The C-terminal domain of the DNA polymerase catalytic subunit regulates the primase and polymerase activities of the human DNA polymerase α-primase complex. J. Biol. Chem. 289, 22021–22034 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizuno T et al. The intrinsically disordered N-terminal region of mouse DNA polymerase α mediates its interaction with POT1a/b at telomeres. Genes Cells 26, 360–380 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Liu H & Naismith JH An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8, 91 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noble AJ et al. Reducing effects of particle adsorption to the air–water interface in cryo-EM. Nat. Methods 15, 793–795 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Punjani A, Zhang H & Fleet DJ Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020). [DOI] [PubMed] [Google Scholar]
- 38.Pettersen EF et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Afonine PV et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Biol. Crystallogr. 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM maps of the human primosome elongation complexes have been deposited to the Electron Microscopy Data Bank (EMDB) under the accession code EMD-27256 (EC-I) and EMD-27258 (EC-II). The corresponding atomic coordinates are deposited in the RSCB Protein Data Bank under the accession code 8D96 (EC-I) and 8D9D (EC-II). The coordinates for the initial models used are available in the PDB under accession codes 5EXR (human p49, p58 and p70) and 4QCL (p180 and template:primer). Source data are provided with this paper.



