Chlamydiota
| Chlamydiota | |
|---|---|
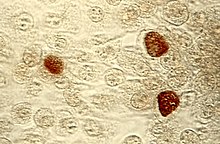
| |
| Chlamydia trachomatis | |
| Scientific classification | |
| Domain: | Bacteria |
| Superphylum: | PVC superphylum |
| Phylum: | Chlamydiota Garrity & Holt 2021[3] |
| Class: | Chlamydiia Horn 2016[1][2] |
| Orders and families | |
| Synonyms | |
| |
The Chlamydiota (synonym Chlamydiae) are a bacterial phylum and class whose members are remarkably diverse, including pathogens of humans and animals, symbionts of ubiquitous protozoa,[4] and marine sediment forms not yet well understood.[5] All of the Chlamydiota that humans have known about for many decades are obligate intracellular bacteria; in 2020 many additional Chlamydiota were discovered in ocean-floor environments, and it is not yet known whether they all have hosts.[5] Historically it was believed that all Chlamydiota had a peptidoglycan-free cell wall, but studies in the 2010s demonstrated a detectable presence of peptidoglycan, as well as other important proteins.[6][7][8][9][10][11]
Among the Chlamydiota, all of the ones long known to science grow only by infecting eukaryotic host cells. They are as small as or smaller than many viruses. They are ovoid in shape and stain Gram-negative. They are dependent on replication inside the host cells; thus, some species are termed obligate intracellular pathogens and others are symbionts of ubiquitous protozoa. Most intracellular Chlamydiota are located in an inclusion body or vacuole. Outside cells, they survive only as an extracellular infectious form.
These Chlamydiota can grow only where their host cells grow, and develop according to a characteristic biphasic developmental cycle.[12][13][14] Therefore, clinically relevant Chlamydiota cannot be propagated in bacterial culture media in the clinical laboratory. They are most successfully isolated while still inside their host cells.
Of various Chlamydiota that cause human disease, the two most important species are Chlamydia pneumoniae, which causes a type of pneumonia, and Chlamydia trachomatis, which causes chlamydia. Chlamydia is the most common bacterial sexually transmitted infection in the United States, and 2.86 million chlamydia infections are reported annually.
History
[edit]Chlamydia-like disease affecting the eyes of people was first described in ancient Chinese and Egyptian manuscripts. A modern description of chlamydia-like organisms was provided by Halberstaedrrter and von Prowazek in 1907.
Chlamydial isolates cultured in the yolk sacs of embryonating eggs were obtained from a human pneumonitis outbreak in the late 1920s and early 1930s, and by the mid-20th century, isolates had been obtained from dozens of vertebrate species. The term chlamydia (a cloak) appeared in the literature in 1945, although other names continued to be used, including Bedsonia, Miyagawanella, ornithosis-, TRIC-, and PLT-agents. In 1956, Chlamydia trachomatis was first cultured by Tang Fei-fan, though they were not yet recognized as bacteria.[15]
Nomenclature
[edit]In 1966, Chlamydiota were recognized as bacteria and the genus Chlamydia was validated.[16] The order Chlamydiales was created by Storz and Page in 1971. The class Chlamydiia was recently validly published.[17][18][19] Between 1989 and 1999, new families, genera, and species were recognized. The phylum Chlamydiae was established in Bergey's Manual of Systematic Bacteriology.[20] By 2006, genetic data for over 350 chlamydial lineages had been reported.[21] Discovery of ocean-floor forms reported in 2020 involves new clades.[5] In 2022 the phylum was renamed Chlamydiota.[3]
Taxonomy and molecular signatures
[edit]The Chlamydiota currently contain eight validly named genera, and 14 genera.[22] The phylum presently consist of two orders (Chlamydiales, Parachlamydiales) and nine families within a single class (Chlamydiia).[17][18] Only four of these families are validly named (Chlamydiaceae, Parachlamydiaceae, Simkaniaceae, Waddliaceae)[23][24] while five are described as families (Clavichlamydiaceae, Criblamydiaceae, Parilichlamydiaceae, Piscichlamydiaceae, and Rhabdochlamydiaceae).[25][26][27]
The Chlamydiales order as recently described contains the families Chlamydiaceae, and the Clavichlamydiaceae, while the new Parachlamydiales order harbors the remaining seven families.[17] This proposal is supported by the observation of two distinct phylogenetic clades that warrant taxonomic ranks above the family level. Molecular signatures in the form of conserved indels (CSIs) and proteins (CSPs) have been found to be uniquely shared by each separate order, providing a means of distinguishing each clade from the other and supporting the view of shared ancestry of the families within each order.[17][28] The distinctness of the two orders is also supported by the fact that no CSIs were found among any other combination of families.
Molecular signatures have also been found that are exclusive for the family Chlamydiaceae.[17][28] The Chlamydiaceae originally consisted of one genus, Chlamydia, but in 1999 was split into two genera, Chlamydophila and Chlamydia. The genera have since 2015 been reunited where species belonging to the genus Chlamydophila have been reclassified as Chlamydia species.[29][30]
However, CSIs and CSPs have been found specifically for Chlamydophila species, supporting their distinctness from Chlamydia, perhaps warranting additional consideration of two separate groupings within the family.[17][28] CSIs and CSPs have also been found that are exclusively shared by all Chlamydia that are further indicative of a lineage independent from Chlamydophila, supporting a means to distinguish Chlamydia species from neighbouring Chlamydophila members.
Phylogenetics
[edit]The Chlamydiota form a unique bacterial evolutionary group that separated from other bacteria about a billion years ago, and can be distinguished by the presence of several CSIs and CSPs.[17][28][31][14] The species from this group can be distinguished from all other bacteria by the presence of conserved indels in a number of proteins and by large numbers of signature proteins that are uniquely present in different Chlamydiae species.[32][33]
Reports have varied as to whether the Chlamydiota are related to the Planctomycetota or Spirochaetota.[34][35] Genome sequencing, however, indicates that 11% of the genes in Protochlamydia amoebophila UWE25 and 4% in the Chlamydiaceae are most similar to chloroplast, plant, and cyanobacterial genes.[14] Cavalier-Smith has postulated that the Chlamydiota fall into the clade Planctobacteria in the larger clade Gracilicutes. However, phylogeny and shared presence of CSIs in proteins that are lineage-specific indicate that the Verrucomicrobiota are the closest free-living relatives of these parasitic organisms.[36] Comparison of ribosomal RNA genes has provided a phylogeny of known strains within Chlamydiota.[21]
Human pathogens and diagnostics
[edit]Three species of Chlamydiota that commonly infect humans are described:
- Chlamydia trachomatis, which causes the eye-disease trachoma and the sexually transmitted infection chlamydia
- Chlamydophila pneumoniae, which causes a form of pneumonia
- Chlamydophila psittaci, which causes psittacosis
The unique physiological status of the Chlamydiota including their biphasic lifecycle and obligation to replicate within a eukaryotic host has enabled the use of DNA analysis for chlamydial diagnostics.[37] Horizontal transfer of genes is evident and complicates this area of research. In one extreme example, two genes encoding histone-like H1 proteins of eukaryotic origin have been found in the prokaryotic genome of C. trachomatis, an obligate intracellular pathogen.
Phylogeny
[edit]| 16S rRNA based LTP_08_2023[38][39][40] | 120 marker proteins based GTDB 08-RS214[41][42][43] | |||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Taxonomy
[edit]The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature (LPSN)[44] and National Center for Biotechnology Information (NCBI)[45]
- "Similichlamydiales" Pallen, Rodriguez-R & Alikhan 2022 [Hat2]
- Family "Piscichlamydiaceae" Horn 2010
- Family "Parilichlamydiaceae" Stride et al. 2013 ["Similichlamydiaceae" Pallen, Rodriguez-R & Alikhan 2022]
- Order Chlamydiales Storz & Page 1971
- Family "Actinochlamydiaceae" Steigen et al. 2013
- Family "Criblamydiaceae" Thomas, Casson & Greub 2006
- Family Chlamydiaceae Rake 1957 ["Clavichlamydiaceae" Horn 2011]
- Family Parachlamydiaceae Everett, Bush & Andersen 1999
- Family Rhabdochlamydiaceae Corsaro et al. 2009
- Family Simkaniaceae Everett, Bush & Andersen 1999
- Family Waddliaceae Rurangirwa et al. 1999
See also
[edit]References
[edit]- ^ Horn M. (2010). "Class I. Chlamydiia class. nov.". In Krieg NR, Staley JT, Brown DR, Hedlund BP, Paster BJ, Ward NL, Ludwig W, Whitman WB. (eds.). Bergey's Manual of Systematic Bacteriology. Vol. 4 (2nd ed.). New York, NY: Springer. p. 844. doi:10.1007/978-0-387-68572-4. ISBN 978-0-387-95042-6.
- ^ Oren A, Garrity GM. (2016). "Validation list no. 170. List of new names and new combinations previously effectively, but not validly, published". Int J Syst Evol Microbiol. 66 (7): 2463–2466. doi:10.1099/ijsem.0.001149. PMID 27530111.
- ^ a b Oren A, Garrity GM (2021). "Valid publication of the names of forty-two phyla of prokaryotes". Int J Syst Evol Microbiol. 71 (10): 5056. doi:10.1099/ijsem.0.005056. PMID 34694987. S2CID 239887308.
- ^ Sixt BS, Siegl A, Müller C, Watzka M, Wultsch A, Tziotis D, et al. (2013). "Metabolic features of Protochlamydia amoebophila elementary bodies—a link between activity and infectivity in Chlamydiae". PLOS Pathogens. 9 (8): e1003553. doi:10.1371/journal.ppat.1003553. PMC 3738481. PMID 23950718.
- ^ a b c Dharamshi JE, Tamarit D, Eme L, Stairs CW, Martijn J, Homa F, et al. (March 2020). "Marine Sediments Illuminate Chlamydiae Diversity and Evolution". Current Biology. 30 (6): 1032–1048.e7. Bibcode:2020CBio...30E1032D. doi:10.1016/j.cub.2020.02.016. PMID 32142706. S2CID 212423997.
- ^ Pilhofer M, Aistleitner K, Biboy J, Gray J, Kuru E, Hall E, et al. (2013-12-02). "Discovery of chlamydial peptidoglycan reveals bacteria with murein sacculi but without FtsZ". Nature Communications. 4 (1): 2856. Bibcode:2013NatCo...4.2856P. doi:10.1038/ncomms3856. PMC 3847603. PMID 24292151.
- ^ Jacquier N, Viollier PH, Greub G (March 2015). "The role of peptidoglycan in chlamydial cell division: towards resolving the chlamydial anomaly". FEMS Microbiology Reviews. 39 (2): 262–275. doi:10.1093/femsre/fuv001. PMID 25670734.
- ^ Malhotra M, Sood S, Mukherjee A, Muralidhar S, Bala M (September 2013). "Genital Chlamydia trachomatis: an update". The Indian Journal of Medical Research. 138 (3): 303–316. PMC 3818592. PMID 24135174.
- ^ Liechti GW, Kuru E, Hall E, Kalinda A, Brun YV, VanNieuwenhze M, Maurelli AT (February 2014). "A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis". Nature. 506 (7489): 507–510. Bibcode:2014Natur.506..507L. doi:10.1038/nature12892. PMC 3997218. PMID 24336210.
- ^ Liechti G, Kuru E, Packiam M, Hsu YP, Tekkam S, Hall E, et al. (May 2016). "Pathogenic Chlamydia Lack a Classical Sacculus but Synthesize a Narrow, Mid-cell Peptidoglycan Ring, Regulated by MreB, for Cell Division". PLOS Pathogens. 12 (5): e1005590. doi:10.1371/journal.ppat.1005590. PMC 4856321. PMID 27144308.
- ^ Gupta RS (August 2011). "Origin of diderm (Gram-negative) bacteria: antibiotic selection pressure rather than endosymbiosis likely led to the evolution of bacterial cells with two membranes". Antonie van Leeuwenhoek. 100 (2): 171–182. doi:10.1007/s10482-011-9616-8. PMC 3133647. PMID 21717204.
- ^ Horn M (2008). "Chlamydiae as symbionts in eukaryotes". Annual Review of Microbiology. 62: 113–131. doi:10.1146/annurev.micro.62.081307.162818. PMID 18473699. S2CID 13405815.
- ^ Abdelrahman YM, Belland RJ (November 2005). "The chlamydial developmental cycle". FEMS Microbiology Reviews. 29 (5): 949–959. doi:10.1016/j.femsre.2005.03.002. PMID 16043254.
- ^ a b c Horn M, Collingro A, Schmitz-Esser S, Beier CL, Purkhold U, Fartmann B, et al. (April 2004). "Illuminating the evolutionary history of Chlamydiae". Science. 304 (5671): 728–730. Bibcode:2004Sci...304..728H. doi:10.1126/science.1096330. PMID 15073324. S2CID 39036549.
- ^ Philip S. Brachman and Elias Abrutyn (2009-07-23). Bacterial Infections of Humans: Epidemiology and Control. ISBN 9780387098425.
- ^ Moulder JW (1966). "The relation of the psittacosis group (Chlamydiae) to bacteria and viruses". Annual Review of Microbiology. 20: 107–130. doi:10.1146/annurev.mi.20.100166.000543. PMID 5330228.
- ^ a b c d e f g Gupta RS, Naushad S, Chokshi C, Griffiths E, Adeolu M (September 2015). "A phylogenomic and molecular markers based analysis of the phylum Chlamydiae: Proposal to divide the class Chlamydiia into two orders, Chlamydiales and Parachlamydiales ord. nov., and emended description of the class Chlamydiia". Antonie van Leeuwenhoek. 108 (3): 765–781. doi:10.1007/s10482-015-0532-1. PMID 26179278. S2CID 17099157.
- ^ a b Oren A, Garrity GM (July 2016). "List of new names and new combinations previously effectively, but not validly, published". International Journal of Systematic and Evolutionary Microbiology. 66 (7): 2463–2466. doi:10.1099/ijsem.0.001149. PMID 27530111.
- ^ Storz J, Page LA (1971). "Taxonomy of the Chlamydiae: reasons for classifying organisms of the genus Chlamydia, family Chlamydiaceae, in a separate order, Chlamydiales ord. nov". International Journal of Systematic Bacteriology. 21 (4): 332–334. doi:10.1099/00207713-21-4-332.
- ^ Garrity GM, Boone DR (2001). Bergey's Manual of Systematic Bacteriology Volume 1: The Archaea and the Deeply Branching and Phototrophic Bacteria (2nd ed.). Springer. ISBN 978-0-387-98771-2.
- ^ a b Everett KD, Thao M, Horn M, Dyszynski GE, Baumann P (July 2005). "Novel chlamydiae in whiteflies and scale insects: endosymbionts 'Candidatus Fritschea bemisiae' strain Falk and 'Candidatus Fritschea eriococci' strain Elm". International Journal of Systematic and Evolutionary Microbiology. 55 (Pt 4): 1581–1587. doi:10.1099/ijs.0.63454-0. PMID 16014485.
- ^ Sayers; et al. "Chlamydiia". National Center for Biotechnology Information (NCBI) taxonomy database. Retrieved 2016-10-24.
- ^ Everett KD, Bush RM, Andersen AA (April 1999). "Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms". International Journal of Systematic Bacteriology. 49 (Pt 2): 415–440. doi:10.1099/00207713-49-2-415. PMID 10319462.
- ^ Rurangirwa FR, Dilbeck PM, Crawford TB, McGuire TC, McElwain TF (April 1999). "Analysis of the 16S rRNA gene of micro-organism WSU 86-1044 from an aborted bovine foetus reveals that it is a member of the order Chlamydiales: proposal of Waddliaceae fam. nov., Waddlia chondrophila gen. nov., sp. nov". International Journal of Systematic Bacteriology. 49 (Pt 2): 577–581. doi:10.1099/00207713-49-2-577. PMID 10319478.
- ^ Thomas V, Casson N, Greub G (December 2006). "Criblamydia sequanensis, a new intracellular Chlamydiales isolated from Seine river water using amoebal co-culture". Environmental Microbiology. 8 (12): 2125–2135. Bibcode:2006EnvMi...8.2125T. doi:10.1111/j.1462-2920.2006.01094.x. PMID 17107554. S2CID 31211875.
- ^ Stride MC, Polkinghorne A, Miller TL, Groff JM, Lapatra SE, Nowak BF (March 2013). "Molecular characterization of "Candidatus Parilichlamydia carangidicola," a novel Chlamydia-like epitheliocystis agent in yellowtail kingfish, Seriola lalandi (Valenciennes), and the proposal of a new family, "Candidatus Parilichlamydiaceae" fam. nov. (order Chlamydiales)". Applied and Environmental Microbiology. 79 (5): 1590–1597. Bibcode:2013ApEnM..79.1590S. doi:10.1128/AEM.02899-12. PMC 3591964. PMID 23275507.
- ^ Kuo C-C, Horn M, Stephens RS (2011) Order I. Chlamydiales. In: Bergey's Manual of Systematic Bacteriology, vol. 4, 2nd ed. pp. 844-845. Eds Krieg N, Staley J, Brown D, Hedlund B, Paster B, Ward N, Ludwig W, Whitman W. Springer-: New York.
- ^ a b c d Griffiths E, Ventresca MS, Gupta RS (January 2006). "BLAST screening of chlamydial genomes to identify signature proteins that are unique for the Chlamydiales, Chlamydiaceae, Chlamydophila and Chlamydia groups of species". BMC Genomics. 7: 14. doi:10.1186/1471-2164-7-14. PMC 1403754. PMID 16436211.
- ^ Sachse K, Bavoil PM, Kaltenboeck B, Stephens RS, Kuo CC, Rosselló-Móra R, Horn M (March 2015). "Emendation of the family Chlamydiaceae: proposal of a single genus, Chlamydia, to include all currently recognized species". Systematic and Applied Microbiology. 38 (2): 99–103. Bibcode:2015SyApM..38...99S. doi:10.1016/j.syapm.2014.12.004. hdl:10261/123714. PMID 25618261.
- ^ Oren A, Garrity GM (2015). "List of new names and new combinations previously effectively, but not validly, published". Int J Syst Evol Microbiol. 65 (7): 2017–2025. doi:10.1099/ijs.0.000317. PMID 28056215.
- ^ Greub G, Raoult D (September 2003). "History of the ADP/ATP-translocase-encoding gene, a parasitism gene transferred from a Chlamydiales ancestor to plants 1 billion years ago". Applied and Environmental Microbiology. 69 (9): 5530–5535. Bibcode:2003ApEnM..69.5530G. doi:10.1128/AEM.69.9.5530-5535.2003. PMC 194985. PMID 12957942.
- ^ Griffiths E, Petrich AK, Gupta RS (August 2005). "Conserved indels in essential proteins that are distinctive characteristics of Chlamydiales and provide novel means for their identification". Microbiology. 151 (Pt 8): 2647–2657. doi:10.1099/mic.0.28057-0. PMID 16079343.
- ^ Gupta RS, Griffiths E (December 2006). "Chlamydiae-specific proteins and indels: novel tools for studies". Trends in Microbiology. 14 (12): 527–535. doi:10.1016/j.tim.2006.10.002. PMID 17049238.
- ^ Ward NL, Rainey FA, Hedlund BP, Staley JT, Ludwig W, Stackebrandt E (November 2000). "Comparative phylogenetic analyses of members of the order Planctomycetales and the division Verrucomicrobia: 23S rRNA gene sequence analysis supports the 16S rRNA gene sequence-derived phylogeny". International Journal of Systematic and Evolutionary Microbiology. 50 (Pt 6): 1965–1972. doi:10.1099/00207713-50-6-1965. PMID 11155969.
- ^ Teeling H, Lombardot T, Bauer M, Ludwig W, Glöckner FO (May 2004). "Evaluation of the phylogenetic position of the planctomycete 'Rhodopirellula baltica' SH 1 by means of concatenated ribosomal protein sequences, DNA-directed RNA polymerase subunit sequences and whole genome trees". International Journal of Systematic and Evolutionary Microbiology. 54 (Pt 3): 791–801. doi:10.1099/ijs.0.02913-0. PMID 15143026.
- ^ Griffiths E, Gupta RS (August 2007). "Phylogeny and shared conserved inserts in proteins provide evidence that Verrucomicrobia are the closest known free-living relatives of chlamydiae". Microbiology. 153 (Pt 8): 2648–2654. doi:10.1099/mic.0.2007/009118-0. PMID 17660429. S2CID 2094762.
- ^ Corsaro D, Greub G (April 2006). "Pathogenic potential of novel Chlamydiae and diagnostic approaches to infections due to these obligate intracellular bacteria". Clinical Microbiology Reviews. 19 (2): 283–297. doi:10.1128/CMR.19.2.283-297.2006. PMC 1471994. PMID 16614250.
- ^ "The LTP". Retrieved 20 November 2023.
- ^ "LTP_all tree in newick format". Retrieved 20 November 2023.
- ^ "LTP_08_2023 Release Notes" (PDF). Retrieved 20 November 2023.
- ^ "GTDB release 08-RS214". Genome Taxonomy Database. Retrieved 10 May 2023.
- ^ "bac120_r214.sp_label". Genome Taxonomy Database. Retrieved 10 May 2023.
- ^ "Taxon History". Genome Taxonomy Database. Retrieved 10 May 2023.
- ^ J.P. Euzéby. "Chlamydiota". List of Prokaryotic names with Standing in Nomenclature (LPSN). Retrieved 2022-09-09.
- ^ Sayers; et al. "Chlamydiae". National Center for Biotechnology Information (NCBI) taxonomy database. Retrieved 2022-09-09.
External links
[edit]- Ward M. "www.chlamydiae.com". University of Southampton.
The comprehensive reference and education wiki on Chlamydia and the Chlamydiales
- Chlamydia Overview

