Abstract
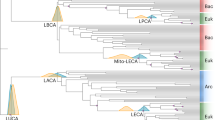
The concept of a last universal common ancestor of all cells (LUCA, or the progenote) is central to the study of early evolution and life's origin, yet information about how and where LUCA lived is lacking. We investigated all clusters and phylogenetic trees for 6.1 million protein coding genes from sequenced prokaryotic genomes in order to reconstruct the microbial ecology of LUCA. Among 286,514 protein clusters, we identified 355 protein families (∼0.1%) that trace to LUCA by phylogenetic criteria. Because these proteins are not universally distributed, they can shed light on LUCA's physiology. Their functions, properties and prosthetic groups depict LUCA as anaerobic, CO2-fixing, H2-dependent with a Wood–Ljungdahl pathway, N2-fixing and thermophilic. LUCA's biochemistry was replete with FeS clusters and radical reaction mechanisms. Its cofactors reveal dependence upon transition metals, flavins, S-adenosyl methionine, coenzyme A, ferredoxin, molybdopterin, corrins and selenium. Its genetic code required nucleoside modifications and S-adenosyl methionine-dependent methylations. The 355 phylogenies identify clostridia and methanogens, whose modern lifestyles resemble that of LUCA, as basal among their respective domains. LUCA inhabited a geochemically active environment rich in H2, CO2 and iron. The data support the theory of an autotrophic origin of life involving the Wood–Ljungdahl pathway in a hydrothermal setting.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Fox, G. E. et al. The phylogeny of prokaryotes. Science 209, 457–463 (1980).
Arndt, N. & Nisbet, E. Processes on the young Earth and the habitats of early life. Annu. Rev. Earth Planet Sci. 40, 521–549 (2012).
Woese, C. The universal ancestor. Proc. Natl Acad. Sci. USA 95, 6854–6859 (1998).
Koonin, E. V. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nature Rev. Microbiol. 1, 127–136 (2003).
Williams, T. A., Foster, P. G., Cox, C. J. & Embley, T. M. An archaeal origin of eukaryotes supports only two primary domains of life. Nature 504, 231–236 (2013).
Raymann, K., Brochier-Armanet, C. & Gribaldo, S. The two-domain tree of life is linked to a new root for the Archaea. Proc. Natl Acad. Sci. USA 112, 6670–6675 (2015).
Ouzounis, C. A., Kunin, V., Darzentas, N. & Goldovsky, L. A minimal estimate for the gene content of the last universal common ancestor—exobiology from a terrestrial perspective. Res. Microbiol. 157, 57–68 (2006).
Kannan, L., Li, H., Rubinstein, B. & Mushegian, A. Models of gene gain and gene loss for probabilistic reconstruction of gene content in the last universal common ancestor of life. Biol. Direct. 8, 32 (2013).
Nelson-Sathi, S. et al. Origins of major archaeal clades correspond to gene acquisitions from bacteria. Nature 517, 77–80 (2015).
Say, R. F. & Fuchs, G. Fructose 1,6-bisphosphate aldolase/phosphatase may be an ancestral gluconeogenic enzyme. Nature 464, 1077–1081 (2010).
Fuchs, G. Alternative pathways of carbon dioxide fixation: insights into the early evolution of life? Annu. Rev. Microbiol. 65, 631–658 (2011).
Baross, J. A. & Hoffman, S. E. Submarine hydrothermal vents and associated gradient environments as sites for the origin and evolution of life. Origins Life Evol. B 15, 327–345 (1985).
Russell, M. J. & Hall, A. J. The emergence of life from iron monosulphide bubbles at a submarine hydrothermal redox and pH front. J. Geol. Soc. Lond. 154, 377–402 (1997).
Buckel, W. & Thauer, R. K. Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation. Biochim. Biophys. Acta 1827, 94–113 (2013).
Schuchmann, K. & Müller, V. Autotrophy at the thermodynamic limit of life: a model for energy conservation in acetogenic bacteria. Nature Rev. Microbiol. 12, 809–821 (2014).
Ferry, J. G. & House, C. H. The step-wise evolution of early life driven by energy conservation. Mol. Biol. Evol. 23, 1286–1292 (2006).
Martin, W. & Russell, M. J. On the origin of biochemistry at an alkaline hydrothermal vent. Phil. Trans. R. Soc. Lond. B 362, 1887–1925 (2007).
Mulkidjanian, A. Y., Galperin, M. Y., Makarova, K. S., Wolf, Y. I. & Koonin, E. V. Evolutionary primacy of sodium bioenergetics. Biol. Direct. 3, 13 (2008).
Lane, N. & Martin, W. F. The origin of membrane bioenergetics. Cell 151, 1406–1416 (2012).
Déclais, A. C., Marsault, J., Confalonieri, F., La Tour de, C. B. & Duguet, M. Reverse gyrase, the two domains intimately cooperate to promote positive supercoiling. J. Biol. Chem. 275, 19498–19504 (2000).
Ragsdale, S. W. Nickel-based enzyme systems. J. Biol. Chem. 284, 18571–18575 (2009).
Broderick, J. B., Duffus, B. R., Duschene, K. S. & Shepard, E. M. Radical S-adenosylmethionine enzymes. Chem. Rev. 114, 4229–4317 (2014).
Eck, R. V. & Dayhoff, M. O. Evolution of the structure of ferredoxin based on living relics of primitive amino acid sequences. Science 152, 363–366 (1966).
Hall, D. O., Cammack, R. & Rao, K. K. Role of ferredoxins in the origin of life and biological evolution. Nature 233, 136–138 (1971).
Böck, A., Forchhammer, K., Heider, J. & Baron, C. Selenoprotein synthesis: an expansion of the genetic code. Trends Biochem. Sci. 16, 463–467 (1991).
Liu, Y. C., Beer, L. L. & Whitman, W. B. Methanogens: a window into ancient sulfur metabolism. Trends Microbiol. 20, 251–258 (2012).
Evans, P. N. et al. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 350, 434–438 (2015).
Lever, M. A. Acetogenesis in the energy-starved deep biosphere—a paradox? Front. Microbiol. 2, 284 (2012).
Schönheit, P., Buckel, W. & Martin, W. F. On the origin of heterotrophy. Trends Microbiol. 24, 12–25 (2016).
Schrenk, M. O., Brazelton, W. J. & Lang, S. Q. Serpentinization, carbon, and deep life. Rev. Mineral. Geochem. 75, 575–606 (2013).
Etiope, G. & Schoell, M. Abiotic gas: atypical, but not rare. Elements 10, 291–296 (2014).
Proskurowski, G. et al. Abiogenic hydrocarbon production at Lost City hydrothermal field. Science 319, 604–607 (2008).
McDermott, J. M., Seewald, J. S., German, C. R. & Sylva, S. P. Pathways for abiotic organic synthesis at submarine hydrothermal fields. Proc. Natl Acad. Sci. USA 112, 7668–7672 (2015).
Chow, C. S., Lamichhane, T. N. & Mahto, S. K. Expanding the nucleotide repertoire of the ribosome with post-transcriptional modifications. ACS Chem. Biol. 2, 610–619 (2007).
Agris, P. F., Vendeix, F. A. P. & Graham, W. D. tRNA's wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 366, 1–13 (2007).
Grosjean, H., Gupta, R., & Maxwell, E. S. in Archaea: New Models for Prokaryotic Biology (ed. Blum, P.) 171–196 (Caister Academic Press, 2008).
Seewald, J. S., Tolotov, M. Y. & McCollom, T. Experimental investigation of single carbon compounds under hydrothermal conditions. Geochim. Cosmochim. Acta 70, 446–460 (2006).
He, C., Tian, G., Liu, Z. & Feng, S. A mild hydrothermal route to fix carbon dioxide to simple carboxylic acids. Org. Lett. 12, 649–651 (2010).
Horita, J. & Berndt, M. Abiogenic methane formation and isotopic fractionation under hydrothermal conditions. Science 285, 1055–1057 (1999).
Amend, J. P. & Shock, E. L. Energetics of amino acid synthesis in hydrothermal ecosystems. Science 281, 1659–1662 (1998).
Amend, J. P., LaRowe, D. E., McCollom, T. M. & Shock, E. L. The energetics of organic synthesis inside and outside the cell. Phil. Trans. R. Soc. Lond. B 368, 20120255 (2013).
Yokoyama, S., Watanabe, K. & Miyazawa, T. Dynamic structures and functions of transfer ribonucleic acids from extreme thermophiles. Adv. Biophys. 23, 115–147 (1987).
Helm, M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 34, 721–733 (2006).
Gottschalk, G. & Thauer, R. K. The Na+-translocating methyltransferase complex from methanogenic archaea. Biochim. Biophys. Acta 1505, 28–36 (2001).
Svetlitchnaia, T., Svetlitchnyi, V., Meyer, O. & Dobbek, H. Structural insights into methyltransfer reactions of a corrinoid iron–sulfur protein involved in acetyl-CoA synthesis. Proc. Natl Acad. Sci. USA 103, 14331–14336 (2006).
Raymond, J. & Segre, D. The effect of oxygen on biochemical networks and the evolution of complex life. Science 311, 1764–1767 (2006).
Dibrova, D. V., Galperin, M. Y. & Mulkidjanian, A. Y. Phylogenomic reconstruction of archaeal fatty acid metabolism. Environ. Microbiol. 16, 907–918 (2014).
Shock, E. L. & Boyd, E. S. Geomicrobiology and microbial geochemistry: principles of geobiochemistry. Elements 11, 389–394 (2015).
Mansy, S. S. et al. Template-directed synthesis of a genetic polymer in a model protocell. Nature 454, 122–125 (2008).
Patel, B. H., Percivalle, C., Ritson, D. J., Duffy, C. D. & Sutherland, J. D. Common origins of RNA, protein and lipid precursors in a cyanosulfidic protometabolism. Nature Chem. 7, 301–307 (2015).
Pruitt, K. D., Tatusova, T., Brown, G. R. & Maglott, D. R. NCBI reference sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 40, D130–D135 (2011).
Enright, A. J., Van Dongen, S. & Ouzounis, C. A. An ancient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30, 1575–1584 (2002).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Rice, P., Longden, I. & Bleasby, A. EMBOSS: The European Molecular Biology open software suite. Trends Genet. 16, 276–277 (2000).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Landan, G. & Graur, D. Heads or tails: a simple reliability check for multiple sequence alignments. Mol. Biol. Evol. 24, 1380–1383 (2007).
Landan, G. & Graur, D. Local reliability measures from sets of co-optimal multiple sequence alignments. Pac. Symp. Biocomput. 13, 15–24 (2008).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Junier, T. & Zdobnov, E. M. The Newick utilities: high-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics 26, 1669–1670 (2010).
Tatusov, R. L., Galperin, M. Y., Natale, D. A. & Koonin, E. V. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28, 33–36 (2000).
Ogata, H. et al. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 27, 29–34 (1999).
Finn, R. D. et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285 (2016).
Machnicka, M. A. et al. MODOMICS: a database of RNA modification pathways—2013 update. Nucleic Acids Res. 41, D262–D267 (2013).
Jühling, F. et al. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 37, D159–D162 (2009).
Humphrey, W., Dalke, A. & Schulten, K. VMD—visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Acknowledgements
The authors thank J. Baross and N. Lane for discussions. The authors acknowledge the Zentrum für Informations- und Medientechnologie (ZIM) of the Heinrich-Heine University for computational support and the European Research Council for funding (ERC AdG 666053 to W.F.M.).
Author information
Authors and Affiliations
Contributions
M.C.W., F.L.S., S.N., M.R. and S.N.-S. performed the bioinformatics analysis. F.L.S. and N.M. carried out the functional classification of the protein families. All authors analysed and discussed the results. W.F.M., F.L.S. and S.N.-S. designed the research. M.C.W., F.L.S., S.N., N.M., S.N.-S. and W.F.M. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary Figures 1-3, Supplementary Tables 1,3-6, legends for Supplementary Tables 2,7-9, Supplementary References (PDF 1698 kb)
Supplementary Table 2
Functional and taxonomic characterization of the 355 protein families potentially present in LUCA using a threshold of 25% global identity (XLSX 95 kb)
Supplementary Table 7
SAM-dependent enzymes (XLSX 32 kb)
Supplementary Table 8
Functional and taxonomic characterization of one-taxa-misplaced protein families (XLSX 34 kb)
Supplementary Table 9
Functional and taxonomic characterization of one-phyla-misplaced protein families (XLSX 34 kb)
Rights and permissions
About this article
Cite this article
Weiss, M., Sousa, F., Mrnjavac, N. et al. The physiology and habitat of the last universal common ancestor. Nat Microbiol 1, 16116 (2016). https://doi.org/10.1038/nmicrobiol.2016.116
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2016.116
This article is cited by
-
Modern analogs for ammonia flux from terrestrial hydrothermal features to the Archean atmosphere
Scientific Reports (2024)
-
The buck stops with spermidine
Nature Chemical Biology (2024)
-
Ion and lipid orchestration of secondary active transport
Nature (2024)
-
Abiotic formation of alkylsulfonic acids in interstellar analog ices and implications for their detection on Ryugu
Nature Communications (2024)
-
Biodiversity of carbapenem-resistant bacteria in clinical samples from the Southwest Amazon region (Rondônia/Brazil)
Scientific Reports (2024)