Geo-Parcel Based Crop Identification by Integrating High Spatial-Temporal Resolution Imagery from Multi-Source Satellite Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Data
2.2.1. Remotely Sensed Imagery
2.2.2. Ancillary Data
2.3. Methods
2.3.1. Geo-Parcels Extraction
2.3.2. Pre-Processing of Medium-Resolution Images
2.3.3. Construction of the Vegetation Index Time-Series
2.3.4. EVI Time Series Smoothing Method
- (1)
- First, the linear interpolation method was applied, to interpolate the missing points along the time profile. The time interval was set as five days, which produces a dense EVI curve.
- (2)
- Then, the S–G filtering method was used to eliminate small unnecessary fluctuations that are caused by system factors. The S–G filter method was applied twice, to enhance the smoothing effects. This procedure aims to highlight key points, namely summit points and bottom points. Here, the S–G parameter window size was set as five, and polynomial degree was set as three.
- (3)
- Based on the slight S–G smoothing process, it became easier to distinguish the summit and bottom points. Several groups of key points were selected. Each group was used to represent a vegetative greenness period. Every group consisted of a summit point (tC) and two bottom points (tL and tR) at tC side, the difference of which was required to be greater than 0.2. A difference that was less than 0.2 was considered to be an abnormal fluctuation, so the corresponding group was discarded. After that, the global time profile was divided into several temporal parts, and each part was from tL to tR.
- (4)
- Furthermore, based on temporal segments, the Gaussian and Polynomial fitting methods were used to smooth the local EVI time profile (Equation (1)), from tL to tR. Figure 5a shows the original data and its fitting result (fcenter) between two bottom points (tL and tR). The Gaussian and Polynomial fitting methods were also conducted between every two neighboring summit points. The original data and their fitted functions are shown in Figure 5b,c. To connect fitted local functions, the function fitting method (Equation (2)) was developed, as shown in Figure 5d.
2.3.5. Calculation of EVI Time-Series Metrics
2.3.6. Crop Identification Using Random Forest
3. Results
3.1. Time Profile Smoothing
3.2. Rotation Frequency Judgement
3.3. Phenological Features Extraction
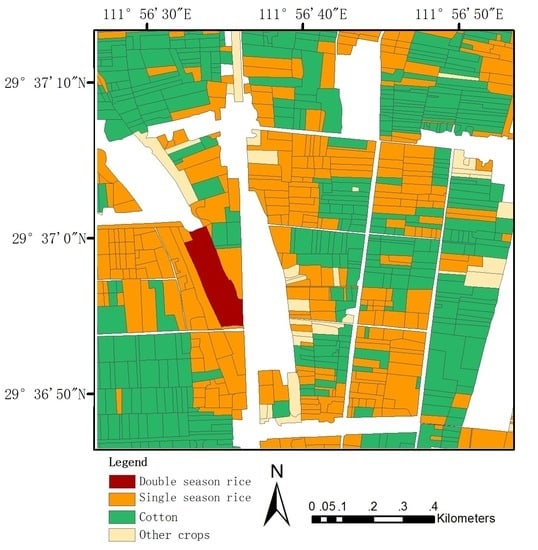
3.4. Crop Classification and Spatial Distribution
4. Discussion
4.1. Comparison of the S–G and HANTS Smoothing Methods
4.2. Classification Accuracy Comparison
4.3. Features Significance Ranking
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Reed, B.C.; Brown, J.F.; Vanderzee, D.; Loveland, T.R.; Merchant, J.W.; Ohlen, D.O. Measuring phenological variability from satellite imagery. J. Veg. Sci. 1994, 5, 703–714. [Google Scholar] [CrossRef]
- Hill, M.J.; Donald, G.E. Estimating spatio-temporal patterns of agricultural productivity in fragmented landscapes using AVHRR NDVI time series. Remote Sens. Environ. 2003, 84, 367–384. [Google Scholar] [CrossRef]
- Sibanda, M.; Murwira, A. The use of multi-temporal MODIS images with ground data to distinguish cotton from maize and sorghum fields in smallholder agricultural landscapes of southern Africa. Int. J. Remote Sens. 2012, 33, 4841–4855. [Google Scholar] [CrossRef]
- Viovy, N.; Arino, O.; Belward, A.S. The Best Index Slope Extraction (BISE): A method for reducing noise in NDVI time-series. Int. J. Remote Sens. 1992, 13, 1585–1590. [Google Scholar] [CrossRef]
- Ma, M.; Veroustraete, F. Reconstructing pathfinder AVHRR land NDVI time-series data for the northwest of China. Adv. Space Res. 2006, 37, 835–840. [Google Scholar] [CrossRef]
- Chen, J.; Jönsson, P.; Tamura, M.; Gu, Z.; Matsushita, B.; Eklundh, L. A simple method for reconstructing a high-quality NDVI time-series data set based on the Savitzky–Golay filter. Remote Sens. Environ. 2004, 91, 332–344. [Google Scholar] [CrossRef]
- Jonsson, P.; Eklundh, L. Seasonality extraction by function fitting to time-series of satellite sensor data. Geosci. Remote Sens. IEEE Trans. 2002, 40, 1824–1832. [Google Scholar] [CrossRef]
- Atzberger, C.; Eilers, P.C. Evaluating the effectiveness of smoothing algorithms in the absence of ground reference measurements. Int. J. Remote Sens. 2011, 32, 3689–3709. [Google Scholar] [CrossRef]
- Zhang, X.; Friedl, M.A.; Schaaf, C.B.; Strahler, A.H.; Hodges, J.C.F.; Gao, F.; Reed, B.C.; Huete, A. Monitoring vegetation phenology using MODIS. Remote Sens. Environ. 2003, 84, 471–475. [Google Scholar] [CrossRef]
- Hird, J.N.; Mcdermid, G.J. Noise reduction of NDVI time series: An empirical comparison of selected techniques. Remote Sens. Environ. 2009, 113, 248–258. [Google Scholar] [CrossRef]
- Turker, M.; Ozdarici, A. Field-based crop classification using SPOT4, SPOT5, IKONOS and QuickBird imagery for agricultural areas: A comparison study. Int. J. Remote Sens. 2011, 32, 9735–9768. [Google Scholar] [CrossRef]
- Immitzer, M.; Atzberger, C.; Koukal, T. Tree species classification with random forest using very high spatial resolution 8-band WordView-2 satellite data. Remote Sens. 2012, 4, 2661–2693. [Google Scholar] [CrossRef]
- Yu, Q.; Gong, P.; Clinton, N.; Biging, G.; Kelly, M.; Schirokauer, D. Object-based detailed vegetation classification with airborne high spatial resolution remote sensing imagery. Photogramm. Eng. Remote Sens. 2006, 72, 799–811. [Google Scholar] [CrossRef]
- Gao, F.; Masek, J.; Schwaller, M.; Hall, F. On the blending of the Landsat and MODIS surface reflectance: Predicting daily Landsat surface reflectance. IEEE Trans. Geosci. Remote Sens. 2006, 44, 2207–2218. [Google Scholar]
- Hilker, T.; Wulder, M.A.; Coops, N.C.; Seitz, N.; White, J.C.; Gao, F.; Masek, J.G.; Stenhouse, G. Generation of dense time series synthetic Landsat data through data blending with MODIS using a spatial and temporal adaptive reflectance fusion model. Remote Sens. Environ. 2009, 113, 1988–1999. [Google Scholar] [CrossRef]
- Xie, D.; Zhang, J.; Zhu, X.; Pan, Y.; Liu, H.; Yuan, Z.; Yun, Y. An improved STARFM with help of an Unmixing-based method to generate high spatial and temporal resolution remote sensing data in complex heterogeneous regions. Sensors 2016, 16, 207. [Google Scholar] [CrossRef] [PubMed]
- Conrad, C.; Fritsch, S.; Zeidler, J.; Rücker, G.; Dech, S. Per-field irrigated crop classification in arid Central Asia using SPOT and ASTER data. Remote Sens. 2010, 2, 1035–1056. [Google Scholar] [CrossRef] [Green Version]
- Singha, M.; Wu, B.; Zhang, M. Object-based paddy rice mapping using HJ-1A/B data and temporal features extracted from time series MODIS NDVI data. Sensors 2016, 17, 10. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Qin, Z.; Zeng, Z. A study on the relative radiometric normalization of multi-sources and multi-temporal remote sensing data. J. Geo-Inf. Sci. 2016, 18, 606–614. [Google Scholar]
- Wardlow, B.D.; Egbert, S.L.; Kastens, J.H. Analysis of time-series MODIS 250m vegetation index data for crop classification in the U.S. Central great plains. Remote Sens. Environ. 2007, 108, 290–310. [Google Scholar] [CrossRef]
- North, H.C.; Pairman, D.; Belliss, S.E.; Mcneill, S.J. Spectral classification of crop groups for land use identification with temporally sparse time-series satellite images. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium, Melbourne, Australia, 21–26 July 2013; pp. 4237–4240. [Google Scholar]
- Demir, B.; Bovolo, F.; Bruzzone, L. Updating land-cover maps by classification of image time series: A novel change-detection-driven transfer learning approach. IEEE Trans. Geosci. Remote Sens. 2013, 51, 300–312. [Google Scholar] [CrossRef]
- Hongjun, L.I.; Zheng, L.; Lei, Y.; Chunqiang, L.I.; Zhou, K. Comparison of NDVI and EVI based on EOS/MODIS data. Prog. Geogr. 2007, 26, 26–32. [Google Scholar]
- Jönsson, P.; Eklundh, L. Timesat—A program for analyzing time-series of satellite sensor data. Comput. Geosci. 2004, 30, 833–845. [Google Scholar] [CrossRef]
- Atkinson, P.M.; Jeganathan, C.; Dash, J.; Atzberger, C. Inter-comparison of four models for smoothing satellite sensor time-series data to estimate vegetation phenology. Remote Sens. Environ. 2012, 123, 400–417. [Google Scholar] [CrossRef]
- Fan, C.; Zheng, B.; Myint, S.W.; Aggarwal, R. Characterizing changes in cropping patterns using sequential Landsat imagery: An adaptive threshold approach and application to Phoenix, Arizona. Int. J. Remote Sens. 2014, 35, 7263–7278. [Google Scholar] [CrossRef]
- Huang, Q.; Qin, Z.; Zeng, Z. Study on the crop classification and planting area estimation at land parcel scale using multisources satellite data. J. Geo-Inf. Sci. 2016, 18, 708–717. [Google Scholar]
- Tucker, C.J.; Elgin, J.H.; McMurtrey, J.E.; Fan, C.J. Monitoring corn and soybean crop development with hand-held radiometer spectral data. Remote Sens. Environ. 1979, 8, 237–248. [Google Scholar] [CrossRef]
- Hao, P.; Zhan, Y.; Wang, L.; Niu, Z.; Shakir, M. Feature selection of time series MODIS data for early crop classification using random forest: A case study in Kansas, USA. Remote Sens. 2015, 7, 5347–5369. [Google Scholar] [CrossRef]
- Breiman, L. Random forest. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]


















| Satellite | Payloads | Bands No. | Spectral Range (μm) | Spatial Resolution (m) | Swath Width (km) | Repetition Cycle (Day) |
|---|---|---|---|---|---|---|
| GF-1 | WFV | 1 | 0.45–0.52 | 16 | 800 (four cameras combined) | 4 |
| 2 | 0.52–0.59 | |||||
| 3 | 0.63–0.69 | |||||
| 4 | 0.77–0.89 |
| Satellite | Sensor | Acquisition Time | Day of Year |
|---|---|---|---|
| GF-1 | WFV2 | 8-02-2016 | 38 |
| GF-1 | WFV1 | 28-03-2016 | 88 |
| GF-1 | WFV3 | 18-04-2016 | 109 |
| GF-1 | WFV4 | 05-05-2016 | 126 |
| GF-1 | WFV1 | 12-05-2016 | 133 |
| GF-1 | WFV1 | 16-05-2016 | 137 |
| GF-1 | WFV2 | 14-06-2016 | 166 |
| GF-1 | WFV1 | 14-06-2016 | 166 |
| GF-1 | WFV4 | 23-06-2016 | 175 |
| GF-1 | WFV2 | 09-07-2016 | 191 |
| GF-1 | WFV3 | 09-07-2016 | 191 |
| GF-1 | WFV1 | 25-07-2016 | 207 |
| GF-1 | WFV1 | 29-07-2016 | 211 |
| GF-1 | WFV3 | 15-08-2016 | 228 |
| GF-1 | WFV3 | 15-08-2016 | 228 |
| GF-1 | WFV3 | 03-10-2016 | 277 |
| GF-1 | WFV1 | 04-11-2016 | 308 |
| GF-1 | WFV4 | 26-11-2016 | 330 |
| GF-1 | WFV4 | 04-12-2016 | 339 |
| GF-1 | WFV1 | 15-12-2016 | 350 |
| Landsat 8 | OLI | 30-07-2016 | 212 |
| Landsat 8 | OLI | 16-09-2016 | 260 |
| All Features | Paddy Rice | Paddy Rice-Cole | Cole-Paddy Rice | Double Season Paddy Rice | Cole-Cotton | Cole-Paddy Rice-Cole | Other Crops | User’s Accuracy |
|---|---|---|---|---|---|---|---|---|
| Paddy Rice | 79 | 0 | 0 | 0 | 0 | 0 | 29 | 0.7315 |
| Paddy Rice-Cole | 0 | 170 | 0 | 0 | 0 | 1 | 0 | 0.9942 |
| Cole-Paddy Rice | 0 | 0 | 218 | 0 | 21 | 1 | 0 | 0.9083 |
| Double Season Paddy Rice | 0 | 0 | 1 | 115 | 2 | 0 | 0 | 0.9746 |
| Cole-Cotton | 0 | 0 | 12 | 0 | 121 | 0 | 0 | 0.9098 |
| Cole-Paddy Rice-Cole | 0 | 0 | 0 | 0 | 0 | 245 | 3 | 0.9880 |
| Other Crops | 3 | 1 | 7 | 0 | 0 | 2 | 202 | 0.9395 |
| Overall Accuracy | 0.9327 | |||||||
| Kappa | 0.9201 |
| Spectral Features | Paddy Rice | Paddy Rice-Cole | Cole-Paddy Rice | Double Season Paddy Rice | Cole-Cotton | Cole-Paddy Rice-Cole | Other Crops | User’s Accuracy |
|---|---|---|---|---|---|---|---|---|
| Paddy Rice | 80 | 2 | 5 | 0 | 0 | 1 | 20 | 0.7407 |
| Paddy Rice-Cole | 60 | 78 | 2 | 0 | 0 | 30 | 1 | 0.4561 |
| Cole-Paddy Rice | 10 | 0 | 201 | 0 | 21 | 8 | 0 | 0.8375 |
| Double Season Paddy Rice | 1 | 0 | 0 | 114 | 0 | 0 | 3 | 0.9661 |
| Cole-Cotton | 1 | 0 | 13 | 0 | 118 | 0 | 1 | 0.8872 |
| Cole-Paddy Rice-Cole | 1 | 1 | 19 | 0 | 4 | 224 | 1 | 0.8960 |
| Other Crops | 13 | 1 | 8 | 0 | 5 | 0 | 188 | 0.8744 |
| Overall Accuracy | 0.8121 | |||||||
| Kappa | 0.7777 |
| Phenological Features | Paddy Rice | Paddy Rice-Cole | Cole-Paddy Rice | Double Season Paddy Rice | Cole-Cotton | Cole-Paddy Rice-Cole | Other Crops | User’s Accuracy |
|---|---|---|---|---|---|---|---|---|
| Paddy Rice | 102 | 0 | 0 | 0 | 0 | 0 | 6 | 0.9444 |
| Paddy Rice-Cole | 0 | 170 | 0 | 0 | 0 | 1 | 0 | 0.9942 |
| Cole-Paddy Rice | 0 | 0 | 168 | 0 | 71 | 1 | 0 | 0.7000 |
| Double Season paddy Rice | 0 | 0 | 0 | 113 | 3 | 2 | 0 | 0.9576 |
| Cole-Cotton | 0 | 0 | 17 | 0 | 116 | 0 | 0 | 0.8722 |
| Cole-Paddy Rice-Cole | 0 | 0 | 0 | 0 | 5 | 243 | 0 | 0.9798 |
| Other Crops | 23 | 0 | 5 | 3 | 0 | 2 | 182 | 0.8465 |
| Overall Accuracy | 0.8873 | |||||||
| Kappa | 0.8672 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Huang, Q.; Wu, W.; Luo, J.; Gao, L.; Dong, W.; Wu, T.; Hu, X. Geo-Parcel Based Crop Identification by Integrating High Spatial-Temporal Resolution Imagery from Multi-Source Satellite Data. Remote Sens. 2017, 9, 1298. https://doi.org/10.3390/rs9121298
Yang Y, Huang Q, Wu W, Luo J, Gao L, Dong W, Wu T, Hu X. Geo-Parcel Based Crop Identification by Integrating High Spatial-Temporal Resolution Imagery from Multi-Source Satellite Data. Remote Sensing. 2017; 9(12):1298. https://doi.org/10.3390/rs9121298
Chicago/Turabian StyleYang, Yingpin, Qiting Huang, Wei Wu, Jiancheng Luo, Lijing Gao, Wen Dong, Tianjun Wu, and Xiaodong Hu. 2017. "Geo-Parcel Based Crop Identification by Integrating High Spatial-Temporal Resolution Imagery from Multi-Source Satellite Data" Remote Sensing 9, no. 12: 1298. https://doi.org/10.3390/rs9121298
APA StyleYang, Y., Huang, Q., Wu, W., Luo, J., Gao, L., Dong, W., Wu, T., & Hu, X. (2017). Geo-Parcel Based Crop Identification by Integrating High Spatial-Temporal Resolution Imagery from Multi-Source Satellite Data. Remote Sensing, 9(12), 1298. https://doi.org/10.3390/rs9121298