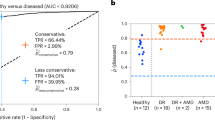
Abstract
In ophthalmologic practice, retinal images are routinely obtained to diagnose and monitor primary eye diseases and systemic conditions affecting the eye, such as diabetic retinopathy. Recent studies have shown that biomarkers on retinal images, for example, retinal blood vessel density or tortuosity, are associated with cardiac function and may identify patients at risk of coronary artery disease. In this work we investigate the use of retinal images, alongside relevant patient metadata, to estimate left ventricular mass and left ventricular end-diastolic volume, and subsequently, predict incident myocardial infarction. We trained a multichannel variational autoencoder and a deep regressor model to estimate left ventricular mass (4.4 (–32.30, 41.1) g) and left ventricular end-diastolic volume (3.02 (–53.45, 59.49) ml) and predict risk of myocardial infarction (AUC = 0.80 ± 0.02, sensitivity = 0.74 ± 0.02, specificity = 0.71 ± 0.03) using just the retinal images and demographic data. Our results indicate that one could identify patients at high risk of future myocardial infarction from retinal imaging available in every optician and eye clinic.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All UKB images and demographic data are available, with restrictions, from UKB. Researchers who use the UKB dataset must first complete the UKB online access management system application form. More information on accessing the UKB dataset can be found in this link: https://www.ukbiobank.ac.uk/enable-your-research/apply-for-access. The AREDS dataset (NCT00000145) is available in the dbGAP repository: https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000001.v3.p1.
Code availability
All algorithms used in this study were developed using libraries and scripts in PyTorch. The source code is publicly available at https://doi.org/10.5281/zenodo.5716142.
References
Roth, G. A. et al. Global, regional, and national burden of cardiovascular diseases for 10 causes, 1990 to 2015. J. Am. College Cardiol. 70, 1–25 (2017).
D’Agostino, R. B. et al. General cardiovascular risk profile for use in primary care. Circulation 117, 743–753 (2008).
Wong, T. Y. et al. Retinal microvascular abnormalities and their relationship with hypertension, cardiovascular disease, and mortality. Survey Ophthalmol. 46 1, 59–80 (2001).
McClintic, B. R., McClintic, J. I., Bisognano, J. D. & Block, R. C. The relationship between retinal microvascular abnormalities and coronary heart disease: a review. Am. J. Med. 123, 374–e1 (2010).
Poplin, R. et al. Predicting cardiovascular risk factors from retinal fundus photographs using deep learning. Nat. Biomed. Eng. 2, 158–164 (2018).
Cheung, C. et al. Quantitative and qualitative retinal microvascular characteristics and blood pressure. J. Hypertens. 27, 1380–1391 (2011).
Tapp, R. J. et al. Associations of retinal microvascular diameters and tortuosity with blood pressure and arterial stiffness. Hypertension 74, 1383–1390 (2019).
Gargeya, R. & Leng, T. Automated identification of diabetic retinopathy using deep learning. Ophthalmology 124, 962–969 (2017).
Qummar, S. et al. A deep learning ensemble approach for diabetic retinopathy detection. IEEE Access 7, 150530–150539 (2019).
Pickhardt, P. J. et al. Automated CT biomarkers for opportunistic prediction of future cardiovascular events and mortality in an asymptomatic screening population: a retrospective cohort study. Lancet Digit. Health 2, e192–e200 (2020).
Petersen, S. E. et al. UK Biobank’s cardiovascular magnetic resonance protocol. J. Cardiovasc. Magn. Reson. 18, 1–7 (2015).
MacGillivray, T. J. et al. Suitability of UK Biobank retinal images for automatic analysis of morphometric properties of the vasculature. PLoS ONE 10, 1–10 (2015).
Fu, H. et al. Evaluation of retinal image quality assessment networks in different color-spaces. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2019 48–56 (Springer, 2019).
Kaggle Diabetic Retinopathy Competition (Kaggle, accessed 19 January 2020); https://www.kaggle.com/c/diabetic-retinopathy-detection/data
Zheng, Q., Delingette, H., Duchateau, N. & Ayache, N. 3D Consistent and robust segmentation of cardiac images by deep learning with spatial propagation. IEEE Trans. Med. Imaging 37, 2137–2148 (2018).
Khaw, K.-T. et al. Association of hemoglobin A1c with cardiovascular disease and mortality in adults: the European prospective investigation into cancer in Norfolk. Annals Intern. Med. 141, 413–420 (2004).
Levitan, E. et al. Hba1c measured in stored erythrocytes and mortality rate among middle-aged and older women. Diabetologia 51, 267–275 (2008).
Gerstein, H. C. et al. The hemoglobin A1c level as a progressive risk factor for cardiovascular death, hospitalization for heart failure, or death in patients with chronic heart failure: an analysis of the Candesartan in heart failure: assessment of reduction in mortality and morbidity (CHARM) program. Arch. Intern. Med. 168, 1699–1704 (2008).
Skali, H. et al. Cardiac structure and function across the glycemic spectrum in elderly men and women free of prevalent heart disease: the atherosclerosis risk in the community study. Circ. Heart Fail. 8, 448–454 (2015).
Petersen, S. E. et al. Reference ranges for cardiac structure and function in cardiovascular magnetic resonance (CMR) imaging in Caucasians from the UK Biobank population cohort. J. Cardiovasc. Magn. Reson. 19, 18 (2017).
Attar, R. et al. 3D Cardiac shape prediction with deep neural networks: simultaneous use of images and patient metadata. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2019 586–594 (Springer, 2019).
The Age-Related Eye Disease Study Sytem Group et al.The Age-Related Eye Disease Study (AREDS): design implications AREDS report number 1. Control. Clin. Trials 20, 573–600 (1999).
The Age-Related Eye Disease Study System Groupet al. The Age-Related Eye Disease Study system for classifying age-related macular degeneration from stereoscopic color fundus photographs: the Age-Related Eye Disease Study report number 6. Am. J. Ophthalmol. 132, 668–681 (2001).
Antelmi, L., Ayache, N., Robert, P. & Lorenzi, M. Sparse multi-channel variational autoencoder for the joint analysis of heterogeneous data. In Proc. 36th International Conference on Machine Learning Vol. 97, 302–311 (PMLR, 2019).
Kingma, D. P. & Welling, M. Auto-encoding variational Bayes. Proc. 2nd International Conference on Learning Representations (ICLR, 2014).
Rezende, D. J., Mohamed, S. & Wierstra, D. Stochastic backpropagation and approximate inference in deep generative models. Proc. 31st International Conference on International Conference on Machine Learning, 32, 1278–1286 (ICML, 2014).
Hotelling, H. Relations between two sets of variates. Biometrika 28, 321–377 (1936).
Haufe, S. et al. On the interpretation of weight vectors of linear models in multivariate neuroimaging. NeuroImage 87, 96–110 (2014).
Johnson, J. M. & Khoshgoftaar, T. M. Survey on deep learning with class imbalance. J. Big Data 6, 27 (2019).
Suinesiaputra, A. et al. Fully-automated left ventricular mass and volume MRI analysis in the UK Biobank population cohort: evaluation of initial results. Int. J. Cardiovasc. Imag. 34, 281–291 (2018).
Attar, R. et al. Quantitative CMR population imaging on 20,000 subjects of the UK Biobank imaging study: LV/RV quantification pipeline and its evaluation. Med. Image Anal. 56, 26–42 (2019).
Keith, N. Some different types of essential hypertension: their course and prognosis. Am. J. Med. Sci. 268, 336–345 (1974).
Bai, W. et al. Automated cardiovascular magnetic resonance image analysis with fully convolutional networks. J. Cardiovasc. Magn. Reson. 20, 65 (2018).
Lin, K. et al. Automated assessment of left ventricular function and mass using heart deformation analysis: initial experience in 160 older adults. Acad. Radiol. 23, 321–325 (2016).
Heusel, M., Ramsauer, H., Unterthiner, T., Nessler, B. & Hochreiter, S. GANs trained by a two time-scale update rule converge to a local Nash equilibrium. Proc. 31st International Conference on Neural Information Processing Systemsm 30, 6629–6640 (NeurIPS, 2017).
Ferris, F. L. et al. Clinical classification of age-related macular degeneration. Ophthalmology 120, 844–851 (2013).
Trinh, M., Kalloniatis, M. & Nivison-Smith, L. Vascular changes in intermediate age-related macular degeneration quantified using optical coherence tomography angiography. Transl. Vis. Sci. Technol. 8, 20 (2019).
Grammer, T. B. et al. Cardiovascular risk algorithms in primary care: results from the detect study. Sci. Rep. 9, 1–12 (2019).
Qureshi, W. T. et al. Impact of replacing the pooled cohort equation with other cardiovascular disease risk scores on atherosclerotic cardiovascular disease risk assessment (from the multi-ethnic study of atherosclerosis [mesa]). Am. J. Cardiol. 118, 691–696 (2016).
Wallisch, C. et al. External validation of two framingham cardiovascular risk equations and the pooled cohort equations: a nationwide registry analysis. Int. J. Cardiol. 283, 165–170 (2019).
Wallisch, C. et al. Re-estimation improved the performance of two framingham cardiovascular risk equations and the pooled cohort equations: a nationwide registry analysis. Sci. Rep. 10, 1–11 (2020).
Acknowledgements
A.F.F. is supported by the Royal Academy of Engineering Chair in Emerging Technologies Scheme (grant no. CiET1819\19), the MedIAN Network (grant no. EP/N026993/1) funded by the Engineering and Physical Sciences Research Council (EPSRC). This work was also supported by the Intramural Research Program of National Library of Medicine and National Eye Institute, National Institutes of Health. This research was also supported by the European Union’s Horizon 2020 InSilc (grant no. 777119) and EPSRC TUSCA (grant no. EP/V04799X/1) programmes. E.D. acknowledges funding from the BHF grant FS/13/71/30378.
Author information
Authors and Affiliations
Contributions
A.D.-P. designed and executed all experiments, conducted all subsequent statistical analyses, and drafted the manuscript. N.R. helped design the experiments, helped with the writing, data interpretation and made substantial revisions and edits of the draft manuscript. R.A. helped design the experiments, contributed to the data analysis and data cleaning. A.S. and Y.Z. contributed to the data analysis. E.L., E.D., C.P.G., R.P.G. and S.P. contributed to the analysis of retinal and cardiac magnetic resonance images and shaped the medical contribution of this work. M.L. contributed to the design and implementation of the mcVAE. Q.C., T.D.L.K., E.A., E.Y.C. and Z.L. contributed to the external validation of the proposed method. A.F.F. conceived the methodology, helped design the experiments and contributed to the writing. All authors contributed to the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Machine Intelligence thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Variance Comparison.
Comparison of the joint latent space variance: Obtained distribution of the latent variables when using the mcVAE on only retinal images and both retina + CMR.
Extended Data Fig. 2 FID.
Distribution of the Frechet Inception Distance (FID) score: Obtained distribution of the FID score on the reconstructed CMR images.
Extended Data Fig. 3 LinearRegression Coefficients.
Coefficients obtained from the logistic regression applied to myocardial infarction prediction using the estimated LVM/LVEDV and demographic data. Chol stands for cholesterol, bmi for body mass index, ads for alcohol daily consumption status, sbpa and dbpa stand for systolic and diastolic blood pressure, respectively.
Extended Data Fig. 4 Bland-Altman plots for different retinal image sizes.
Bland-Altman plots, correlation plots, and ROC curves for estimated LVM/LVEDV and MI prediction using different retinal image sizes. The solid line represents the logistic regression, and the dotted line represents the identity line.
Extended Data Fig. 5 Bland-Altman plots for different dataset sizes.
Bland-Altman plots, correlation plots, and ROC curves for estimated LVM/LVEDV and MI prediction using different dataset sizes. The solid line represents the logistic regression, and the dotted line represents the identity line.
Extended Data Fig. 6 Bland-Altman plots for estimated LVM/LVEDV using only retinal images.
Estimation of LVM and LVEDV using retinal images only or demographic data only: Bland-Altman and correlation plots for estimated LVM/LVEDV using only retinal images (upper plots) and only demographic data (lower plots). The solid line represents the logistic regression, and the dotted line represents the identity line.
Extended Data Fig. 7 ROC curves for MI prediction using different demographic data.
ROC curves for MI prediction using different demographic data: An eye clinic (that is age, gender). Accuracy: 0.71 ± 0.01, Sensitivity: 0.74 ± 0.03, Specificity: 0.73 ± 0.06, Precision: 0.68 ± 0.03, and F1 Score: 0.71 ± 0.01. ROC curves for MI prediction using demographic data that may be available at a cardiology department (that is Age, Gender, BMI, Diastolic BP, Systolic BP, HbA1c scores, Glucose, Cholesterol, Smoking and Drinking status). Accuracy: 0.72 ± 0.03, Sensitivity: 0.74 ± 0.02, Specificity: 0.70 ± 0.05, Precision: 0.70 ± 0.05, and F1 Score: 0.72 ± 0.03. The solid line represents the logistic regression, and the dotted line represents the identity line.
Extended Data Fig. 8 Coefficients and ROC curves for MI prediction obtained from the AREDS datasets.
Coefficients and ROC curves for MI prediction obtained from the AREDS datasets: (a) Coefficients obtained from the logistic regression applied to myocardial infarction prediction using the AREDS demographic data (sex, diastolic blood pressure (dbpa), systolic blood pressure (sbpa), smoking status (ss), body mass index (bmi) and age), and (b) ROC curve obtained for 10-fold cross-validation for MI predictions using only demographic data available in the AREDS dataset, and (c) ROC curve obtained for MI predictions using a logistic regression model trained on the UK Biobank and evaluated on the AREDS dataset. The solid line represents the logistic regression, and the dotted line represents the identity line.
Supplementary information
Supplementary Information
Supplementary Figs. 1–8, Tables 1–4 and Methods.
Rights and permissions
About this article
Cite this article
Diaz-Pinto, A., Ravikumar, N., Attar, R. et al. Predicting myocardial infarction through retinal scans and minimal personal information. Nat Mach Intell 4, 55–61 (2022). https://doi.org/10.1038/s42256-021-00427-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-021-00427-7
This article is cited by
-
Prognostic potentials of AI in ophthalmology: systemic disease forecasting via retinal imaging
Eye and Vision (2024)
-
Evaluating imaging repeatability of fully self-service fundus photography within a community-based eye disease screening setting
BioMedical Engineering OnLine (2024)
-
AI-integrated ocular imaging for predicting cardiovascular disease: advancements and future outlook
Eye (2024)
-
Oculomics: A Crusade Against the Four Horsemen of Chronic Disease
Ophthalmology and Therapy (2024)
-
Retinal imaging for the assessment of stroke risk: a systematic review
Journal of Neurology (2024)