Abstract
Fragment-Based Drug Design (FBDD) plays a pivotal role in the field of drug discovery and development. The construction of high-quality fragment libraries is a critical step in FBDD. Conventional fragmentation approaches often rely on rigid rules and chemical intuition, limiting their adaptability to diverse molecular structures. The rapid development of Artificial Intelligence (AI) technology offers a transformative opportunity to rethink traditional methods. Here, we present DigFrag, a digital fragmentation method that highlights important substructures by focusing locally within the molecular graph. In addition, we feed the fragments segmented by machine intelligence and human expertise into the deep generative model to compare the preference for data from different sources. Experimental results show that the structural diversity of fragments segmented by DigFrag is higher, and more desirable compounds are generated based on these fragments. These results also demonstrate that data generated based on AI methods may be more suitable for AI models. Moreover, a user-friendly platform called MolFrag (https://dpai.ccnu.edu.cn/MolFrag/) is developed based on various fragmentation techniques to support molecular segmentation.
Similar content being viewed by others
Introduction
Fragment-based drug design (FBDD) is the process of screening for favorable fragments and combining them to create drug molecules1,2. Compared to traditional high-throughput screening methods, FBDD allows the exploration of a much broader chemical space and the discovery of active compounds with a higher hit rate3. A high-quality fragment library is a prerequisite for FBDD. Typically, the segmentation of a molecule in accordance with predefined guidelines stands as a highly effective means of obtaining these structural fragments, and is of paramount importance in the field of FBDD4. However, it is noteworthy that few innovative studies on fragmentation methods have been reported. Conventional fragmentation methods, exemplified by the likes of RECAP5 and BRICS6, predominantly hinge on the principles of retrosynthesis, where a set of rules govern the breaking of chemical bonds, often emphasizing the selective cleavage of acyclic bonds. As eloquently elucidated by Diao et al., this restriction has its rationale, but it greatly reduces the novelty of the obtained fragments, especially for macrocyclic compounds7,8. Furthermore, these methods do not allow a reasonable selection of fragment size. To elaborate, the diminution of fragment size to an extreme extent may inadvertently disrupt the pharmacophores, while an excessively large fragment may not efficiently extract smaller substituents9.
Over the past decade, artificial intelligence (AI) technologies have achieved great success in many fields. With the increasing abundance of biomedical data, many AI-driven predictive models have been introduced into the field of drug discovery and development, including convolutional neural networks (CNNs)10, recurrent neural networks (RNNs)11, and graph neural networks (GNNs)12. In particular, GNNs consider the input molecules as graphs with attributes, and obtain graph embeddings through the three steps of message passing, aggregation, and updating13. Studies have shown that the attention mechanism serves to improve both the efficiency and accuracy of the model by directing its focus towards feature information that is more relevant to the given task14,15. Combining attention mechanisms with GNNs can better reveal the relative importance of structural fragments within a compound.
Typically, chemists with extensive expertise segment molecules based on chemical rules. In contrast, machine intelligence-oriented perspective, while lacking domain knowledge, provide a distinctive paradigm by segmenting molecules based on mathematical logic (i.e., statistics). Through a comparative analysis of these fragmentation methods, our investigation illuminates that AI-based models segments unique fragments that are feasible in the virtual world but may not be applicable in the real world. This discrepancy between reality and virtuality underscores the intrinsic advantage of AI-based methods, as they enable the exploration of uncharted territories and overcome the limitations of traditional methods. Consequently, the potential of AI to transcend the boundaries of established chemical paradigms becomes manifest, opening frontiers in the field of molecular segmentation.
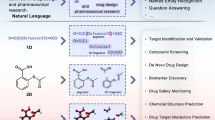
On this basis, we propose DigFrag, a digital fragmentation method based on the graph attention mechanism, which gets rid of the constraints of traditional chemical rules and segments drug/pesticide molecules with attention weights (i.e., contribution to the prediction task) oriented to ultimately obtain the drug/pesticide-like fragments from the perspective of machine intelligence rather than human expertise. Comparison results show that the structural diversity of the fragments segmented by DigFrag is higher. In addition, the application of these fragments to deep generative models yields desirable compounds, highlighting the potential of DigFrag in FBDD. The scheme of this work is depicted in Fig. 1. We believe that DigFrag is a more applicable fragmentation method for AI drug discovery.
Results
Comparison of the property distributions of segmented fragments
Prior to comparison, we trained the models to segment drug/pesticide fragments. The results of the five-fold cross-validation are presented in Supplementary Table 1. It can be seen that these models demonstrated excellent and robust performance across the three key metrics, with accuracy, area under the curve (AUC), and Matthews correlation coefficient (MCC) exceeding 0.90, 0.96, and 0.80, respectively.
Next, we conducted an in-depth evaluation of the fragments obtained by the proposed method (DigFrag), the conventional methods (RECAP5 and BRICS6), and the latest method (MacFrag8), focusing on the property distributions as a critical metric of comparison. As shown in Table 1 and Supplementary Fig. 1, for drug fragments, the DigFrag fragments were much closer to the BRICS fragments in terms of property distributions. For example, the differences in molecular weight and the number of hydrogen bond acceptors between the two groups of fragments were not significant. Interestingly, we found that the DigFrag fragments had a significantly higher number of rotatable bonds compared to the BRICS fragments, which may be attributed to their unique cleavage of cyclic bonds. However, for pesticide fragments, the properties of the DigFrag fragments were distributed in lower regions (Table 2 and Supplementary Fig. 2). Taking molecular weight as an example, the average molecular weight of DigFrag fragments was 137.07, signifying a substantial reduction in comparison with the other three fragments. It should be noted that all properties of the DigFrag fragments showed a similar trend, whether they were drugs or pesticides. In addition, we calculated the number of fragments obtained following the segmentation of each drug/pesticide molecule by the four methods. As shown in Supplementary Fig. 3, none of these methods, except MacFrag, segmented the molecule into more than 20 fragments. In terms of distributions, DigFrag displayed greater similarity to BRICS, and both methods tended to segment smaller fragments.
Comparison of the structural diversity of segmented fragments
We then performed a rigorous analysis of the four methods, with a primary focus on the structural diversity of the segmented fragments. As shown in Supplementary Table 2, our findings demonstrated a remarkable disparity. Specifically, for both drugs and pesticides, the number of duplicates between the fragments segmented by the DigFrag method and the other three methods was low (9.97–21.37% for drugs and 8.94–15.20% for pesticides). In contrast, the fragments segmented by the MacFrag method almost covered the BRICS fragments (96.42% for drugs and 99.05% for pesticides) and the RECAP fragments (76.59% for drugs and 76.31% for pesticides). This suggests that the MacFrag method is only an extension of the conventional methods, while the proposed DigFrag method is able to obtain more unique fragments. In addition, we counted the occurrence numbers of each fragment in the four fragmentation methods and visualized their spatial distribution using the t-distributed Stochastic Neighbor Embedding (t-SNE) algorithm. As illustrated in Supplementary Figs. 4–5, over 75% of the drug/pesticide fragments occurred in only one method. The proportion of drug/pesticide fragments shared by two methods (mainly MacFrag) was approximately 20%, while less than 5% of the remaining fragments occurred in three or four methods. It is noteworthy that despite the small number of fragments shared by the four methods, these had a more dispersed distribution in chemical space compared to the fragments shared by the three methods.
To provide a more comprehensive insight into the assessment of structural diversity, we employed the fragment cluster ratio as a quantitative measure, which provides a more intuitive reflection of the overall structural diversity within the fragment sets. As shown in Fig. 2 and Supplementary Tables 3–4, the DigFrag method consistently yielded higher fragment cluster ratios than other methods. In particular, for the most commonly used similarity thresholds (0.4 and 0.6), the fragments segmented by the DigFrag method exhibited greater structural diversity.
Comparison of the qualities of generated molecules
Subsequently, we conducted a comparative analysis of generative models utilizing distinct sets of fragments on the MOSES benchmarking platform16. As shown in Tables 3–4, the drug molecules generated by the MacFrag-based model achieved optimal results in terms of uniqueness and diversity, but the Filters score was significantly lower than that of the other models, which suggests that these compounds are unsatisfactory in terms of safety. In contrast, the Filters score of the drug molecules generated by the DigFrag-based model was 0.828, indicating the improved safety of compounds modified by digital fragments compared to other fragments. We speculate that this observed superiority may be attributed to the inherent capacity of deep learning models to incorporate considerations of toxicity risk, stability, and other pertinent factors during the fragmentation process, which is not possible with the other three methods. Similarly, except for a slightly higher proportion of duplications, the pesticide molecules generated by the DigFrag-based model achieved superior performance in terms of SMILES validity, novelty, scaffold diversity, and structure alerts. Moreover, an examination of the average Quantitative Estimation of Drug-likeness (QED)17 and Synthetic Accessibility (SA)18 revealed that drug/pesticide molecules generated by the DigFrag-based model outperformed those generated by the other models, as illustrated in Fig. 3. To quantitatively compare the similarity between the property distributions of the generated molecules and the MOSES dataset, the Wasserstein-1 distance provided by the platform was utilized. As shown in Supplementary Tables 5–6, the drug/pesticide molecules generated by the DigFrag model exhibited the highest similarity in terms of three property distributions (molecular weight, QED, and SA). Taken together, these results demonstrate that FBDD employing DigFrag fragments yields molecules of higher quality. Furthermore, the findings suggest a preference within AI-based models for AI-sourced data, reinforcing the potential advantages of leveraging AI in molecular design.
Binding affinity calculation of generated molecules to specific targets
Considering that the DeepFMPO architecture does not incorporate information about the target during the optimization of molecular structures, we performed molecular docking to assess the binding strength of the modified drug/pesticide molecules to the dopamine receptor D2 (DRD2)/4-hydroxyphenylpyruvate dioxygenase (HPPD). As anticipated, marginal disparities in binding affinity were observed between the drug/pesticide molecules generated by the four methods. Specifically, the binding free energy ranges of the generated compounds were −9.16 to −9.36 Kcal/mol (DRD2) and −8.37 to −8.53 Kcal/mol (HPPD), respectively (Supplementary Fig. 6). It is noteworthy that, despite the lack of a pronounced advantage for the drug/pesticide molecules generated by the DigFrag method, they manifested an acceptable level of binding to the corresponding targets, indicating the preservation of desired properties and high affinities.
Binding mode analysis of selected molecules to specific targets
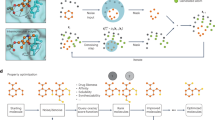
Through the application of refined filters, we identified 24 desirable drug molecules and 20 desirable pesticide molecules that satisfy QED values greater than 0.75, SA values less than 3, and binding free energies less than the positive drug Domperidone (−10.7 Kcal/mol)19/positive pesticide Mesotrione (−8.4 Kcal/mol)20. The chemical structures of these selected molecules are presented in Supplementary Figs. 7–8. Subsequently, an in-depth analysis of the interaction between several desirable drug/pesticide molecules and the corresponding targets was performed. As shown in Figs. 4–5, all drug ligands were effectively docked into the active pocket of DRD2 and formed hydrogen bonds with amino acid residues SER-193, TYR-416, THR-119, VAL-115, TRP-413, and GLU-95. Similarly, all pesticide ligands were stably bound to HPPD through hydrogen bonding interactions with amino acid residues SER-65, SER328, ASP-211, ASP-387, TYR-88, TYR-103, and LEU-78. Intriguingly, the generated compounds exhibited a distinct binding mode compared to the positive drug, suggesting a potential pharmacological mechanism that warrants further exploration in future investigations.
Web server construction and application
Although several fragmentation methods have been reported, no easy-to-use web server is available. Therefore, an innovative platform called MolFrag (https://dpai.ccnu.edu.cn/MolFrag/) was constructed that seamlessly combines four molecular fragmentation approaches, ensuring accessibility to researchers with varying levels of expertise (Fig. 6). We believe that the application of MolFrag streamlines the process of molecular segmentation, making it an invaluable resource for the scientific community.
A Different fragmentation methods are available to users to segment the uploaded molecules. If users have no data, MolFrag provides FDA-approved drugs and commercial pesticides. B In the jobs page, users can view the fragment information obtained after segmentation of the molecules, including physicochemical property, drug/pesticide-like evaluation and synthetic accessibility.
Discussion
In recent years, the field of drug discovery and development has been profoundly revolutionized by advances in AI technology10,11,12. In this work, we introduced DigFrag, an intuitive fragmentation method that segments molecular structures based on AI-driven models, thereby circumventing the constraints inherent in traditional bond-breaking rules. We comprehensively compared the fragments obtained by machine intelligence with those obtained by human expertise. The results demonstrate the superior efficacy of DigFrag in the segmentation of diverse molecular fragments. Furthermore, we fed two initial sets of compounds with the four sets of drug/pesticide fragments into an existing deep generative model. The outcomes of this experiment exhibited the propensity of the DigFrag-derived model to generate molecules characterized by desired properties, thereby substantiating that AI-based models may be more compatible with AI-sourced data.
When we review conventional fragmentation methods, it becomes evident that they are limited in certain contexts. Chemists have successfully refined these methods through extensive experience and domain knowledge. However, in the evolving scientific environment, we are faced with increasingly complex molecular systems. Therefore, we want to explore how we can leverage the power of AI to drive the evolution of molecular fragmentation methods. In addition, we focus not only on the limitations of traditional methods, but also on the differences in data adaptability between machine intelligence and human expertise.
In the FBDD study, we integrated the fragments segmented by the four methods (BRICS, RECAP, MacFrag and DigFrag) into the DeepFMPO framework. To our surprise, AI-based models appeared to be more effective in utilizing the data derived from AI-based methods, and the generated molecules exhibited remarkable excellence in most of the evaluation metrics. Moreover, conventional fragmentation methods tended to ignore target-related information, while DigFrag was able to incorporate this information, which facilitated the segmentation of the same molecular entities into distinct fragments, thereby leading to more personalized molecular design.
However, it is imperative to acknowledge that there is discernible room for improvement within the proposed method. Firstly, an important direction for future research lies in conducting a comprehensive investigation to explore the underlying reasons why data from AI-based methods exhibit increased compatibility with AI-based models. Secondly, recognizing the limitations associated with the two-dimensional graph, which inherently neglects the geometric space of compounds, we aspire to segment molecules based on the three-dimensional graph. This endeavor is expected to yield richer conformational information about the fragments. Furthermore, since the development of fragment-based deep generative models is still in its infancy, we are committed to developing more deep learning frameworks for specific targets, thus expanding the practical application of this method.
Overall, our findings demonstrate that data generated based on AI methods may be more applicable to AI models. We strongly believe that this idea establishes a versatile research paradigm with far-reaching implications in various scientific domains (e.g., materials). For future work, we will strive to extend this idea to a broader range of subtasks, thereby generating more expansive data available for AI-based model exploitation.
Methods
Dataset curation
The modeling dataset utilized in this study was sourced from our self-constructed database called PADFrag21. This database primarily contains 1652 United States Food and Drug Administration (FDA)-approved drugs cataloged in DrugBank and 1259 commercial pesticides listed in Alan Wood. To improve data consistency, structurally non-standard compounds were systematically excluded. The dataset was then partitioned into distinct subsets for the purposes of training, validation, and testing in an 8:1:1 ratio.
AI-based fragmentation method
Construction of molecular graph
In the field of drug discovery and development, GNN-based architectures have been shown to achieve excellent predictive performance for various subtasks12,22. The molecular graph, serving as the input representation of GNNs, is defined as G = (V, E), where V stands for nodes (atoms) and E stands for connecting edges (chemical bonds). In this work, the featurization of these elements was executed using the AttentiveFPAtomFeaturizer and AttentiveFPBondFeaturizer modules of the open-source toolkit DGL-LifeSci23 (Supplementary Table 7).
Model architecture
In this study, we employed the AttentiveFP framework introduced by Xiong et al. to capture the structural information of molecules24. This architecture represents a feature extraction network based on the graph attention mechanism. Specifically, the original molecular graph is initially fed into a series of attention layers to obtain individual atomic embeddings. Subsequently, these embeddings are combined into a unified vector, commonly referred to as a super node. Finally, the entire molecular embedding is obtained through multiple attention layers. It is noteworthy that the attention layer consists of two essential components: the aggregation module (Eqs. 1–3) based on the attention mechanism and the update module (Eqs. 4–5) based on the gated recurrent unit (GRU).
where \({h}_{v}\) and \({h}_{u}\) represent the vector of node \(v\) and the neighbor node \(u\), and \(W\) is a trainable weight matrix.
where \(k\) represent iteration number, and \(M\) is the message function.
Segmentation of molecules
Considering the latent relationship between structural fragments and drug/pesticide-likeness, DigFrag was used to leverage the graph attention mechanism to segment the molecular graph into subgraphs of different sizes (i.e., structural fragments). Consecutively, the fragments with relatively diminished relevance were filtered out by setting an attention weight threshold, while the fragments closely related to the drug/pesticide-likeness were retained. Moreover, the conventional fragmentation method (BRICS and RECAP) was implemented in RDKit, and the latest fragmentation method (MacFrag) was implemented through the source code released by the authors.
Comparative analysis of the fragments obtained by the four methods
In the present study, we conducted a comparative assessment of segmented fragments from two aspects. At the structural level, we generated the corresponding Morgan fingerprint for each fragment using the GetMorganFingerprintAsBitVect module of RDKit. For a more rigorous quantitative evaluation, we executed Butina clustering based on the Tanimoto Coefficient25 for distinct groups of fragments using the BulkTanimotoSimilarity and ClusterData modules of RDKit and then calculated the fragment cluster ratio, which is denoted as the ratio of fragment clusters to the total number of fragments. Higher values of this metric indicate greater diversity in the structural landscape. At the property level, we used the open-source software PaDEL-Descriptor26 to calculate several attributes related to the drug/pesticide-likeness rules, including molecular weight, LogP, topological polar surface area, number of hydrogen bond donors, number of hydrogen bond acceptors, number of rotatable bonds27,28. It should be noted that the calculation of fingerprints and attributes did not consider the breaking position of the fragments, mainly due to the fact that these representations are not relevant to the broken bond information.
Fragment-based molecular generation
To further elucidate the influence of digital and traditional fragments on fragment-based deep generative models, we undertook an investigation utilizing the DeepFMPO29 architecture developed by Ståhl et al. with a specific focus on its application in de novo drug design. DeepFMPO is an actor-critic reinforcement learning model that acquires the ability to generate compounds with desired properties by replacing fragments within a compound. Therefore, the generation of molecules using DeepFMPO requires a balanced binary tree library and an initial set of lead molecules. In this study, the former are the fragments segmented by different methods, and the latter are the molecules collected from the ChEMBL database30 that did not fulfill the sweet spot criteria (561 molecules for DRD2 and 221 molecules for HPPD). The model architecture used a bidirectional long short-term memory (LSTM) network, with the number of neurons in the hidden layer being 128, 64, and 32, respectively. The learning rate and batch size were set to 1e-4 and 512, respectively. Model training was executed until 4000 epochs were reached, after which all molecules generated in the last 100 epochs were compiled and a random selection of 1000 molecules was sampled as representatives for subsequent analysis. Performance evaluation of each model was conducted on the MOSES benchmarking platform16.
Molecular docking of the generated molecules
Docking studies were performed to assess the protein-ligand binding affinity of representative molecules with DRD2 (drug) and HPPD (pesticide). The three-dimensional structures of the targets were obtained from the Protein Data Bank database31 (ID: 6CM432 and 5YWG33) and subsequently subjected to dehydration, hydrogenation, and charge calculation using AutoDockTools software. The three-dimensional structures of representative drug/pesticide molecules were constructed using ChemOffice software34. Binding free energy calculations were performed using Autodock Vina software35. Furthermore, the visualization of protein-ligand interactions was achieved through the utilization of PyMOL software36.
Data availability
The data required to reproduce our results are available at https://doi.org/10.5281/zenodo.13997755. The numerical source data of main figures are available in Supplementary Data 1. All other data are available from the corresponding authors upon reasonable request.
Code availability
The source code implemented in this study is available at https://github.com/yang1rq/MolFrag.
References
Bian, Y. & Xie, X. S. Computational fragment-based drug design: current trends, strategies, and applications. Aaps. J. 20, 59 (2018).
Fattori, D. Molecular recognition: the fragment approach in lead generation. Drug. Discov. Today 9, 229–238 (2004).
Sheng, C. & Zhang, W. Fragment informatics and computational fragment-based drug design: an overview and update. Med. Res. Rev. 33, 554–598 (2013).
Erlanson, D. A., Davis, B. J. & Jahnke, W. Fragment-based drug discovery: advancing fragments in the absence of crystal structures. Cell. Chem. Biol. 26, 9–15 (2019).
Lewell, X. Q., Judd, D. B., Watson, S. P. & Hann, M. M. RECAP–retrosynthetic combinatorial analysis procedure: a powerful new technique for identifying privileged molecular fragments with useful applications in combinatorial chemistry. J. Chem. Inf. Comput. Sci. 38, 511–522 (1998).
Degen, J., Wegscheid-Gerlach, C., Zaliani, A. & Rarey, M. On the art of compiling and using ‘drug-like’ chemical fragment spaces. ChemMedChem 3, 1503–1507 (2008).
Cummings, M. D. & Sekharan, S. Structure-based macrocycle design in small-molecule drug discovery and simple metrics to identify opportunities for macrocyclization of small-molecule ligands. J. Med. Chem. 62, 6843–6853 (2019).
Diao, Y., Hu, F., Shen, Z. & Li, H. MacFrag: segmenting large-scale molecules to obtain diverse fragments with high qualities. Bioinformatics 39, btad012 (2023).
Cramer, J., Sager, C. P. & Ernst, B. Hydroxyl groups in synthetic and natural-product-derived therapeutics: a perspective on a common functional group. J. Med. Chem. 62, 8915–8930 (2019).
Zheng, L., Fan, J. & Mu, Y. OnionNet: a multiple-layer intermolecular-contact-based convolutional neural network for protein-ligand binding affinity prediction. ACS Omega 4, 15956–15965 (2019).
Segler, M. H. S., Kogej, T., Tyrchan, C. & Waller, M. P. Generating focused molecule libraries for drug discovery with recurrent neural networks. ACS Cent. Sci. 4, 120–131 (2018).
Yang, K. et al. Analyzing learned molecular representations for property prediction. J. Chem. Inf. Model. 59, 3370–3388 (2019).
Zhang, Z., Guan, J. & Zhou, S. FraGAT: a fragment-oriented multi-scale graph attention model for molecular property prediction. Bioinformatics 37, 2981–2987 (2021).
Cai, H., Zhang, H., Zhao, D., Wu, J. & Wang, L. FP-GNN: a versatile deep learning architecture for enhanced molecular property prediction. Brief. Bioinform. 23, bbac408 (2022).
Wu, Z. et al. Mining toxicity information from large amounts of toxicity data. J. Med. Chem. 64, 6924–6936 (2021).
Polykovskiy, D. et al. Molecular sets (MOSES): a benchmarking platform for molecular generation models. Front. Pharmacol. 11, 565644 (2020).
Bickerton, G. R., Paolini, G. V., Besnard, J., Muresan, S. & Hopkins, A. L. Quantifying the chemical beauty of drugs. Nat. Chem. 4, 90–98 (2012).
Ertl, P. & Schuffenhauer, A. Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions. J. Cheminform. 1, 8 (2009).
Xiang, Z., Ma, H., Mou, Y. & Xu, C. F. Association between polymorphism of dopamine D2 receptor genes and therapeutic effect of domperidone in functional dyspepsia. Turk. J. Gastroenterol. 26, 1–5 (2015).
Yu, X. H. et al. Discovery and development of 4-hydroxyphenylpyruvate dioxygenase as a novel crop fungicide target. J. Agric. Food Chem. 71, 19396–19407 (2023).
Yang, J. F. et al. PADFrag: a database built for the exploration of bioactive fragment space for drug discovery. J. Chem. Inf. Model. 58, 1725–1730 (2018).
Feinberg, E. N. et al. PotentialNet for molecular property prediction. ACS Cent. Sci. 4, 1520–1530 (2018).
Li, M. et al. DGL-LifeSci: an open-source toolkit for deep learning on graphs in life science. ACS Omega 6, 27233–27238 (2021).
Xiong, Z. et al. Pushing the boundaries of molecular representation for drug discovery with the graph attention mechanism. J. Med. Chem. 63, 8749–8760 (2020).
Zhang, B., Vogt, M., Maggiora, G. M. & Bajorath, J. Design of chemical space networks using a Tanimoto similarity variant based upon maximum common substructures. J. Comput. Aided Mol. Des. 29, 937–950 (2015).
Yap, C. W. PaDEL-descriptor: an open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 32, 1466–1474 (2011).
Lipinski, C. A., Lombardo, F., Dominy, B. W. & Feeney, P. J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46, 3–26 (2001).
Tice, C. M. Selecting the right compounds for screening: does Lipinski’s Rule of 5 for pharmaceuticals apply to agrochemicals? Pest. Manag. Sci. 57, 3–16 (2001).
Ståhl, N., Falkman, G., Karlsson, A., Mathiason, G. & Boström, J. Deep reinforcement learning for multiparameter optimization in de novo drug design. J. Chem. Inf. Model. 59, 3166–3176 (2019).
Mendez, D. et al. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 47, D930–D940 (2019).
Goodsell, D. S. et al. RCSB Protein Data Bank: enabling biomedical research and drug discovery. Protein Sci. 29, 52–65 (2020).
Wang, S. et al. Structure of the D2 dopamine receptor bound to the atypical antipsychotic drug risperidone. Nature 555, 269–273 (2018).
Lin, H. Y. et al. Molecular insights into the mechanism of 4-hydroxyphenylpyruvate dioxygenase inhibition: enzyme kinetics, X-ray crystallography and computational simulations. FEBS J. 286, 975–990 (2019).
Buntrock, R. E. ChemOffice ultra 7.0. J. Chem. Inf. Comput. Sci. 42, 1505–1506 (2002).
Trott, O. & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010).
Yuan, S., Chan, H. C. S. & Hu, Z. Using PyMOL as a platform for computational drug design. Wires. Comput. Mol. Sci. 7, e1298 (2017).
Acknowledgements
This work is supported by the National Key Research and Development Program of China (2023YFD1700500) and the National Natural Science Foundation of China (No. 21837001).
Author information
Authors and Affiliations
Contributions
F.W. and G.Y. conceived the project. R.Y. and H.Z. performed the computational experiments. F.W. and R.Y. collected and analyzed the data. R.Y. wrote the manuscript. F.W. and G.Y. provided evaluation and suggestions.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Chemistry thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, R., Zhou, H., Wang, F. et al. DigFrag as a digital fragmentation method used for artificial intelligence-based drug design. Commun Chem 7, 258 (2024). https://doi.org/10.1038/s42004-024-01346-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42004-024-01346-5