Abstract
Substantial follicle remodelling during the regression phase of the hair growth cycle is coordinated by the contraction of the dermal sheath smooth muscle, but how dermal-sheath-generated forces are regulated is unclear. Here, we identify spatiotemporally controlled endothelin signalling—a potent vasoconstriction-regulating pathway—as the key activating mechanism of dermal sheath contraction. Pharmacological blocking or genetic ablation of both endothelin receptors, ETA and ETB, impedes dermal sheath contraction and halts follicle regression. Epithelial progenitors at the club hair–epithelial strand bottleneck produce the endothelin ligand ET-1, which is required for follicle regression. ET signalling in dermal sheath cells and downstream contraction is dynamically regulated by cytoplasmic Ca2+ levels through cell membrane and sarcoplasmic reticulum calcium channels. Together, these findings illuminate an epithelial–mesenchymal interaction paradigm in which progenitors—destined to undergo programmed cell death—control the contraction of the surrounding sheath smooth muscle to orchestrate homeostatic tissue regression and reorganization for the next stem cell activation and regeneration cycle.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Raw and analysed RNA-seq data supporting the findings of this study are available at the Gene Expression Omnibus (GEO) repository under accession code GSE215133. Previously published RNA-seq data that were reanalysed here are available at the GEO (GSE77197 and GSE136996). All other data supporting the findings of this study are available from the corresponding author on reasonable request. Source data are provided with this paper.
References
Jones, D. L. & Wagers, A. J. No place like home: anatomy and function of the stem cell niche. Nat. Rev. Mol. Cell Biol. 9, 11–21 (2008).
Gonzales, K. A. U. & Fuchs, E. Skin and its regenerative powers: an alliance between stem cells and their niche. Dev. Cell 43, 387–401 (2017).
Fuchs, E. & Blau, H. M. Tissue stem cells: architects of their niches. Cell Stem Cell 27, 532–556 (2020).
Sennett, R. & Rendl, M. Mesenchymal–epithelial interactions during hair follicle morphogenesis and cycling. Semin. Cell Dev. Biol. 23, 917–927 (2012).
Rezza, A., Sennett, R. & Rendl, M. Adult stem cell niches: cellular and molecular components. Curr. Top. Dev. Biol. 107, 333–372 (2014).
Kretzschmar, K. & Clevers, H. Wnt/beta-catenin signaling in adult mammalian epithelial stem cells. Dev. Biol. 428, 273–282 (2017).
Chacon-Martinez, C. A., Koester, J. & Wickstrom, S. A. Signaling in the stem cell niche: regulating cell fate, function and plasticity. Development 145, dev165399 (2018).
Vining, K. H. & Mooney, D. J. Mechanical forces direct stem cell behaviour in development and regeneration. Nat. Rev. Mol. Cell Biol. 18, 728–742 (2017).
Pathak, M. M. et al. Stretch-activated ion channel Piezo1 directs lineage choice in human neural stem cells. Proc. Natl Acad. Sci. USA 111, 16148–16153 (2014).
Aragona, M. et al. Mechanisms of stretch-mediated skin expansion at single-cell resolution. Nature 584, 268–273 (2020).
Scadden, D. T. The stem-cell niche as an entity of action. Nature 441, 1075–1079 (2006).
Scadden, D. T. Nice neighborhood: emerging concepts of the stem cell niche. Cell 157, 41–50 (2014).
Cotsarelis, G., Sun, T. T. & Lavker, R. M. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell 61, 1329–1337 (1990).
Blanpain, C., Lowry, W. E., Geoghegan, A., Polak, L. & Fuchs, E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell 118, 635–648 (2004).
Morris, R. J. et al. Capturing and profiling adult hair follicle stem cells. Nat. Biotechnol. 22, 411–417 (2004).
Tumbar, T. et al. Defining the epithelial stem cell niche in skin. Science 303, 359–363 (2004).
Morgan, B. A. The dermal papilla: an instructive niche for epithelial stem and progenitor cells in development and regeneration of the hair follicle. Cold Spring Harb. Perspect. Med. 4, a015180 (2014).
Hsu, Y. C., Pasolli, H. A. & Fuchs, E. Dynamics between stem cells, niche, and progeny in the hair follicle. Cell 144, 92–105 (2011).
Zhang, Y. V., Cheong, J., Ciapurin, N., McDermitt, D. J. & Tumbar, T. Distinct self-renewal and differentiation phases in the niche of infrequently dividing hair follicle stem cells. Cell Stem Cell 5, 267–278 (2009).
Clavel, C. et al. Sox2 in the dermal papilla niche controls hair growth by fine-tuning BMP signaling in differentiating hair shaft progenitors. Dev. Cell 23, 981–994 (2012).
Harshuk-Shabso, S., Dressler, H., Niehrs, C., Aamar, E. & Enshell-Seijffers, D. Fgf and Wnt signaling interaction in the mesenchymal niche regulates the murine hair cycle clock. Nat. Commun. 11, 5114 (2020).
Yang, H., Adam, R. C., Ge, Y., Hua, Z. L. & Fuchs, E. Epithelial–mesenchymal micro-niches govern stem cell lineage choices. Cell 169, 483–496 (2017).
Mesa, K. R. et al. Niche-induced cell death and epithelial phagocytosis regulate hair follicle stem cell pool. Nature 522, 94–97 (2015).
Paus, R. & Foitzik, K. In search of the ‘hair cycle clock’: a guided tour. Differentiation 72, 489–511 (2004).
Rompolas, P. et al. Live imaging of stem cell and progeny behaviour in physiological hair-follicle regeneration. Nature 487, 496–499 (2012).
Oshimori, N. & Fuchs, E. Paracrine TGF-β signaling counterbalances BMP-mediated repression in hair follicle stem cell activation. Cell Stem Cell 10, 63–75 (2012).
Greco, V. et al. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell 4, 155–169 (2009).
Rompolas, P., Mesa, K. R. & Greco, V. Spatial organization within a niche as a determinant of stem-cell fate. Nature 502, 513–518 (2013).
Heitman, N. et al. Dermal sheath contraction powers stem cell niche relocation during hair cycle regression. Science 367, 161–166 (2020).
Martino, P., Heitman, N. & Rendl, M. The dermal sheath: an emerging component of the hair follicle stem cell niche. Exp. Dermatol. 30, 512–521 (2020).
Hébert, J., Rosenquist, T., Götz, J. & Martin, G. FGF5 as a regulator of the hair growth cycle: evidence from targeted and spontaneous mutations. Cell 78, 1017–1025 (1994).
Kuo, I. Y. & Ehrlich, B. E. Signaling in muscle contraction. Cold Spring Harb. Perspect. Biol. 7, a006023 (2015).
Grisanti, L. et al. Tbx18 targets dermal condensates for labeling, isolation, and gene ablation during embryonic hair follicle formation. J. Invest. Dermatol. 133, 344–353 (2013).
Rendl, M., Lewis, L. & Fuchs, E. Molecular dissection of mesenchymal-epithelial interactions in the hair follicle. PLoS Biol. 3, e331 (2005).
Rezza, A. et al. Signaling networks among stem cell precursors, transit-amplifying progenitors, and their niche in developing hair follicles. Cell Rep. 14, 3001–3018 (2016).
Sennett, R. et al. An integrated transcriptome atlas of embryonic hair follicle progenitors, their niche, and the developing skin. Dev. Cell 34, 577–591 (2015).
Sumner, M. J., Cannon, T. R., Mundin, J. W., White, D. G. & Watts, I. S. Endothelin ETA and ETB receptors mediate vascular smooth muscle contraction. Br. J. Pharmacol. 107, 858–860 (1992).
Guan, Z., VanBeusecum, J. P. & Inscho, E. W. Endothelin and the renal microcirculation. Semin. Nephrol. 35, 145–155 (2015).
Nava, E. & Llorens, S. The paracrine control of vascular motion. A historical perspective. Pharmacol. Res. 113, 125–145 (2016).
Fisher, S. A. Vascular smooth muscle phenotypic diversity and function. Physiol. Genom. 42A, 169–187 (2010).
Rahmani, W. et al. Hair follicle dermal stem cells regenerate the dermal sheath, repopulate the dermal papilla, and modulate hair type. Dev. Cell 31, 543–558 (2014).
Kedzierski, R. M. et al. Cardiomyocyte-specific endothelin A receptor knockout mice have normal cardiac function and an unaltered hypertrophic response to angiotensin II and isoproterenol. Mol. Cell. Biol. 23, 8226–8232 (2003).
Barton, M. & Yanagisawa, M. Endothelin: 30 years from discovery to therapy. Hypertension 74, 1232–1265 (2019).
Davenport, A. P. et al. Endothelin. Pharmacol. Rev. 68, 357–418 (2016).
Bagnall, A. J. et al. Deletion of endothelial cell endothelin B receptors does not affect blood pressure or sensitivity to salt. Hypertension 48, 286–293 (2006).
Neylon, C. B. Vascular biology of endothelin signal transduction. Clin. Exp. Pharm. Physiol. 26, 149–153 (1999).
Zhang, Y. V., White, B. S., Shalloway, D. I. & Tumbar, T. Stem cell dynamics in mouse hair follicles: a story from cell division counting and single cell lineage tracing. Cell Cycle 9, 1504–1510 (2010).
Shohet, R. V. et al. Mice with cardiomyocyte-specific disruption of the endothelin-1 gene are resistant to hyperthyroid cardiac hypertrophy. Proc. Natl Acad. Sci. USA 101, 2088–2093 (2004).
Ghosh, D. et al. Calcium channels in vascular smooth muscle. Adv. Pharmacol. 78, 49–87 (2017).
Stow, L. R., Jacobs, M. E., Wingo, C. S. & Cain, B. D. Endothelin-1 gene regulation. FASEB J. 25, 16–28 (2011).
Maguire, J. J. & Davenport, A. P. ETA receptor-mediated constrictor responses to endothelin peptides in human blood vessels in vitro. Br. J. Pharmacol. 115, 191–197 (1995).
Ling, L., Maguire, J. J. & Davenport, A. P. Endothelin-2, the forgotten isoform: emerging role in the cardiovascular system, ovarian development, immunology and cancer. Br. J. Pharmacol. 168, 283–295 (2013).
Cacioppo, J. A., Koo, Y., Lin, P. C., Gal, A. & Ko, C. Generation and characterization of an endothelin-2 iCre mouse. Genesis 53, 245–256 (2015).
Li, K. N. et al. Skin vasculature and hair follicle cross-talking associated with stem cell activation and tissue homeostasis. eLife 8, e45977 (2019).
Ge, Y. et al. Combined knockout of collecting duct endothelin A and B receptors causes hypertension and sodium retention. Am. J. Physiol. Ren. Physiol. 295, F1635–F1640 (2008).
Cacioppo, J. A. et al. Granulosa cell endothelin-2 expression is fundamental for ovulatory follicle rupture. Sci. Rep. 7, 817 (2017).
Ko, C. et al. Endothelin-2 in ovarian follicle rupture. Endocrinology 147, 1770–1779 (2006).
Ishimoto, S. et al. Role of endothelin receptor signalling in squamous cell carcinoma. Int. J. Oncol. 40, 1011–1019 (2012).
Grimshaw, M. J. Endothelins and hypoxia-inducible factor in cancer. Endocr. Relat. Cancer 14, 233–244 (2007).
Mai, H. Q. et al. Therapeutic targeting of the endothelin a receptor in human nasopharyngeal carcinoma. Cancer Sci. 97, 1388–1395 (2006).
Gehart, H. & Clevers, H. Tales from the crypt: new insights into intestinal stem cells. Nat. Rev. Gastroenterol. Hepatol. 16, 19–34 (2019).
Peng, T. et al. Hedgehog actively maintains adult lung quiescence and regulates repair and regeneration. Nature 526, 578–582 (2015).
Henry, S. P. et al. Generation of aggrecan-CreERT2 knockin mice for inducible Cre activity in adult cartilage. Genesis 47, 805–814 (2009).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010).
Li, L. & Ginty, D. D. The structure and organization of lanceolate mechanosensory complexes at mouse hair follicles. eLife 3, e01901 (2014).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 14, 128 (2013).
Acknowledgements
We thank Sarah Millar for discussions and comments on the manuscript; David Pollock and Ilse Daehn for sharing the Ednra-, Ednrb- and Edn1-targeted mouse lines and Elena Ezhkova for the K14-creERT2 line; Jill Gregory for support with figure illustrations; and the personnel at the ISMMS Flow Cytometry, Microscopy, Genomics and Mouse Genetics CoREs for technical assistance. The ISMMS Microscopy CoRE was supported by NIH Shared Instrumentation grant IS10RR026639. N.H. was supported by training grant T32GM007280 from NIH/NIGMS; and T32HHD075735 from NIH/NIDCR and F30AR070639 from NIH/NIAMS. N.S. was supported by fellowship of the Training Program in Stem Cell Research from the New York State Department of Health (NYSTEM-C32561GG). M.Y. was supported by the World Premier International Research Center Initiative (MEXT), JSPS KAKENHI (17H06095 and 22H04918) and AMED (JP21zf0127005). M.R. was supported by grants from NIH/NIAMS (R01AR071047, R01AR073259, R01AR077593 and R01AR079475) and New York State Department of Health (NYSTEM-C32561GG).
Author information
Authors and Affiliations
Contributions
P.M., R.S., N.H. and M.R. designed the experiments and the overall study. P.M. and M.R. wrote the manuscript. P.M., R.S., N.H., A.B., N.S. and L.G. performed the experiments. D.K. and M.Y. assisted with data analysis and writing the manuscript. M.R. supervised the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks Cédric Blanpain, Anthony Davenport and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Validation of DS, DP and DF isolation strategy during the growth-to-regression transition.
a, Immunofluorescence for pH3 and CASP3* on P16 Tbx18H2BGFP;Crabp1-GFP; Lef1-RFP reporter mouse back skin (n = 2 mice). P13 anagen back skin served as a positive control for pH3. In the hair bulb, proliferation is decreasing and apoptotic cells are beginning to appear, indicating the growth-to-regression transition. Scale bars, 50 µm. b, Immunofluorescence for aSMA, PDGFRA and ITGA8 on P16 Tbx18H2BGFP;Crabp1-GFP;Lef1-RFP reporter mouse back skin (n = 2 mice). Scale bars, 50 µm. c, Immunofluorescence for ITGA8, aSMA, and GFP on P16 Tbx18H2BGFP;Crabp1-GFP;Lef1-RFP reporter mouse back skin, demonstrating colocalization in the dermal sheath (n = 2 mice). Scale bars, 50 µm. d, Immunofluorescence for GFP and RFP on fresh frozen and fixed P16 Tbx18H2BGFP;Crabp1-GFP;Lef1-RFP reporter mouse back skin, both demonstrating colocalization in the DP (n = 2 mice). Scale bars, 50 µm.
Extended Data Fig. 2 Analysis of DP, DS, and DF transcriptomes at the growth-to-regression transition.
a, Principal components analysis of P16 DS, DP, and DF bulk RNA-sequencing data. b, DF, DP and DS RPKM expression levels of Wif1, Acta2 and Lum at regression onset (n = 2, data are mean with individual data points). c, Gene ontology terms enriched in the DS population. ‘Muscle contraction’ is the top enriched GO term for the DS. P < 0.05, Fisher’s exact test. d, Transcription factors, adhesion/ECM molecules, ligands, receptors, and enzymes that are part of the gene signatures of the DS, DP and DF populations. e, Expression of endothelin receptors across cell types from previously published transcriptome data at P520,32(n = 2, data are mean with individual data points).
Extended Data Fig. 3 Endothelin signalling contracts DS cells in a dose-dependent manner.
a, Endothelin-1 (ET-1) triggers DS contraction and surface area reduction in DS cells grown on Matrigel in a dose-dependent manner. Scale bars, 50 µm. b, Quantification of cell surface areas during contraction time course (0, 30, and 60 minutes). 24, 20, 21, 18 and 19 cells for control, 100 pM, 1 nM, 10 nM, 100 nM and 1uM ET-1, respectively; n = 3 independent experiments. **P = 0.001 (30 min) and 0.002 (60 min) for 1 nM ET-1. ***P = 0.00027 (30 min) and 8.65 × 10−5 (60 min) for 10 nM ET-1, 1.24 × 10−5 (30 min) and 2.5 × 10−5 (60 min) for 100 nM ET-1, and 2.53 × 10−5 (30 min) and 1.14 × 10−5 (60 min) for 1 µM ET-1. Data are mean ± s.d. and statistical significance was determined by one-way ANOVA with post-hoc Tukey’s HSD for multiple comparison testing.
Extended Data Fig. 4 Endothelin signalling functionally contracts DS cells independent of proliferation.
a, Endothelin-1 (ET-1) triggers contraction and surface area reduction in DS cells grown on matrigel, both in the absence or presence of proliferation inhibition with Mitomycin-C. Scale bars, 50 µm. b, Quantification of cell surface areas during contraction time course. N = 19, 20, 19 cells for control, ET-1, Mitomycin C + ET-1, respectively, from two independent experiments. **P = 0.0015 (5 min) for Mitomycin C + ET-1; ***P = 0.0002 (5 min), 0.0001 (10 min), 1.42 × 10−5 (20 mins) and 1.12 × 10−5 (30 mins) for ET-1; 0.0002 (10 min), 3.59 × 10−5 (20 min) and 61.90 × 10−5 (30 min) for Mitomycin C + ET-1. Data are mean ± s.d. and statistical significance was determined by one-way ANOVA with post-hoc Tukey’s HSD for multiple comparison testing.
Extended Data Fig. 5 Topical application of endothelin receptor antagonists impedes HF regression with concentrated and local effects.
a, Schematic of the experimental design for topical application of endothelin receptor antagonists BQ123 and BQ788 (‘BQ’) or vehicle control (DMSO), harvesting of the back skins, and imaging strategy. b, Whole-mount immunofluorescence for K14 from the centre of the application area (Zone 1). Most follicles failed to regress in regions treated with BQ123 + BQ788. Scale bars, 50 µm. c, Whole-mount immunofluorescence for K14, LEF1, and DAPI showing a stalled follicle from Zone 1 and a regressed follicle from Zone 3 of BQ123 + BQ788 treated back skin. Scale bars, 50 µm. d, Whole-mount immunofluorescence for K14 at the edge (Zone 2) and just outside of (Zone 3) the application area, demonstrating a progressive decline in stalled follicles towards the periphery. Scale bars, 50 µm. e, Quantification of stalled HFs from each of the three zones. 1200 follicles per zone for vehicle control; 938, 841, and 1087 follicles for Zone 1, 2, and 3, respectively, in BQ123 + BQ788 treated regions; n = 4 mice). *P = 0.0228 (Zone 3), ***P = 1.69 × 10−10 (Zone 1) and 2.42 × 10−7 (Zone 2), unpaired two-tailed Student’s t-test. Data are mean ± s.d. with individual data points.
Extended Data Fig. 6 Topical application of endothelin receptor antagonists impedes follicle regression during the second hair cycle.
a, Schematic of the experimental design for topical application of endothelin receptor antagonists BQ123 and BQ788 (‘BQ’) or vehicle control (DMSO), harvesting of the back skins, and imaging strategy. b, Whole-mount immunofluorescence for K14 and DAPI in DMSO (vehicle control) and BQ123 + BQ788 treated back skin. Scale bars, 200 µm. c, Quantification of stalled HFs observed in DMSO (vehicle control) and BQ123 + BQ788 treated back skins. 924 follicles for DMSO and 544 follicles for BQ123 + BQ788, n = 3 mice. **P = 0.0031, unpaired two-tailed Student’s t-test. Data are mean ± s.d. with individual data points. d, DS-specific Ednra ablation at P17. Low magnification image of the follicle from the high magnification image in Fig. 3b. e, DS-specific Ednrb ablation at P17. Low magnification image of the follicle from the high magnification image in Fig. 3E. Scale bars, 50 µm.
Extended Data Fig. 7 Stalling follicles, but not regressing follicles, in dcKO back skin lack both ETA and ETB receptors.
a, Schematic of DS-specific Ednra and Ednrb double conditional genetic ablation (dcKO) in the skin. b, Immunofluorescence for K14 and both ETA/ETB in control P17 back skins. Endothelin receptors are expressed in the DS. c, Immunofluorescence for K14 and both ETA/ETB in DS-specific dcKO P17 back skin. Only stalling follicles lack both receptors ETA and ETB in the DS. Scale bars, 50 µm. n = 4 mice. Green arrows = presence of ETA and/or ETB in the DS. White arrowheads = lack of both ETA and ETB in the DS.
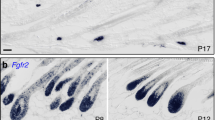
Extended Data Fig. 8 Patterns of endothelin ligand expression during regression.
a, Edn3 mRNA expression across cell types from previously published transcriptome data at P520,32(n = 2, data are mean with individual data points). b, Edn1 smFISH in a follicle transitioning from early-to-mid regression (n = 3 mice). While only very diffuse expression of Edn1 is detectable throughout most of the ORS, the progenitors located in the bottleneck region of the follicle exhibited very high focal expression of Edn1, corresponding to the site of known DS contraction. Scale bars, 50 µm. c, Fire LUT conversion of ET-1 immunofluorescence signal from Fig. 5d highlights the high focal expression of ET-1 in the bottleneck region of regressing follicles. White arrows indicate the bottleneck region. d, Schematic of ET-1 expression during different stages of regression.
Extended Data Fig. 9 ET-1 ablation in ORS progenitors by mid-regression.
Immunofluorescence for K14, ET-1 and CASP3* in P17 back skins from Edn1 cKO and control (n = 4 mice per condition). A marked reduction in both ET-1 and CASP3* in the K14+ ORS progenitors of the HFs in cKO back skins was observed compared to control back skins. Scale bars, 50 µm. b, Insets showing K14 and ET-1 colocalization in regressing HF from control back skin, with high focal expression of ET-1 in the bottleneck region. In Edn1 cKO back skins, some HFs exhibit complete ET-1 ablation while other HFs only exhibit partial ET-1 ablation in the K14+ ORS progenitors. Scale bars, 50 µm.
Extended Data Fig. 10 ET-1-induced DS contraction depends on dynamically regulated cytoplasmic Ca2+.
a, Images of Fluo8 levels in DS cells from Fig. 7b including intermediate timepoints (5, 10, 20 mins) during the 30-minute ET-1 or vehicle control (PBS) exposure; n = 2 independent experiments. Scale bars, 50 µm. b, Images of tdT-labelled DS cells at 0, 5, 10, 20 and 30 minutes of ET-1 or vehicle control (PBS) exposure following a 1 h preincubation in calcium channel blockers (50 µM NNC 55-0396 + 50 µM Diltiazem + 10 µM Ryanodine + 10 µM Xestospongin C), ML7 (200 µM), or vehicle control (DMSO); n = 2 independent experiments. Endothelin-mediated DS contraction is abrogated by either blocking of calcium channels or by inhibition of MLCK activity. Scale bars, 50 µm.
Supplementary information
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Martino, P., Sunkara, R., Heitman, N. et al. Progenitor-derived endothelin controls dermal sheath contraction for hair follicle regression. Nat Cell Biol 25, 222–234 (2023). https://doi.org/10.1038/s41556-022-01065-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-022-01065-w