Abstract
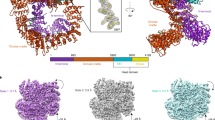
A complex of two proteins, Xrcc4 and DNA ligase IV, plays a fundamental role in DNA non-homologous end joining (NHEJ), a cellular function required for double-strand break repair and V(D)J recombination. Here we report the crystal structure of human Xrcc4 bound to a polypeptide that corresponds to the DNA ligase IV sequence linking its two BRCA1 C-terminal (BRCT) domains. In the complex, a single ligase chain binds asymmetrically to an Xrcc4 dimer. The helical tails of Xrcc4 undergo a substantial conformational change relative to the uncomplexed protein, forming a coiled coil that unwinds upon ligase binding, leading to a flat interaction surface. A buried network of charged hydrogen bonds surrounded by extensive hydrophobic contacts explains the observed tightness of the interaction. The strong conservation of residues at the interface between the two proteins provides evidence that the observed mode of interaction has been maintained in NHEJ throughout evolution.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Critchlow, S.E. & Jackson, S.P. Trends Biochem. Sci. 23, 394–398 (1998).
Jeggo, P.A. Adv. Genet. 38, 185–218 (1998).
Grawunder, U. & Harfst, E. Curr. Opin. Immunol. 13, 186–194 (2001).
Critchlow, S.E., Bowater, R.P. & Jackson, S.P. Curr. Biol. 7, 588–958 (1997).
Grawunder, U. et al. Nature 388, 492–495 (1997).
Li, Z. et al. Cell 83, 1079–1089 (1995).
Gao, Y. et al. Cell 95, 891–902 (1998).
Frank, K.M. et al. Mol. Cell 5, 993–1002 (2000).
Gao, Y. et al. Nature 404, 897–900 (2000).
Barnes, D.E., Stamp, G., Rosewell, I., Denzel, A. & Lindhal, T. Curr. Biol. 8, 1395–1398 (1998).
Junop, M.S. et al. EMBO J. 19, 5962–5970 (2000).
Mizuta, R., Cheng, H.L., Gao, Y. & Alt, F.W. Int. Immunol. 9, 1607–1613 (1997).
Tomkinson, A.E. & Mackey, Z.B. Mutat. Res. 407, 1–9 (1998).
Bork, P. et al. FASEB J. 11, 68–76 (1997).
Grawunder, U., Zimmer, D. & Leiber, M.R. Curr. Biol. 8, 873–876 (1998).
Sibanda, B.L., Blundell, T.L. & Thornton, J.M. J. Mol. Biol. 206, 759–777 (1989).
Modesti, M., Hesse, J.E. & Gellert, M. EMBO J. 18, 2008–2018 (1999).
Keller, W., Konig, P. & Richmond, T.J. J. Mol. Biol. 254, 657–667 (1995).
Gajiwala, K.S. et al. Nature 403, 916–921. (2000).
Otwinowski, Z. & Minor, W. Methods Enzymol. 276, 307–326 (1997).
Miller, R., Gallo, S.M., Khalak, H.G. & Weeks, C.M. J. Appl. Crystallogr. 27, 613–621 (1994).
La Fortelle, E. & Bricogne, G. Methods Enzymol. 276, 472–494 (1997).
Cowtan, K. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography 31, 34–38 (1994).
Brünger, A. et al. Acta Crystallogr. D 54, 905–921 (1998).
Kraulis, P.J. J. Appl. Crystallogr. 24, 946–950 (1991).
Merrit, E.A. & Bacon, D.J. Methods Enzymol. 277, 505–524 (1997).
Acknowledgements
We would like to thank G. Robbins for early work on Xrcc4, and E. Gordon, S. McSweeney, G. Leonard and E. Mitchell for excellent technical assistance at the ESRF beamlines. This research was supported by grants from the Wellcome Trust.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sibanda, B., Critchlow, S., Begun, J. et al. Crystal structure of an Xrcc4–DNA ligase IV complex. Nat Struct Mol Biol 8, 1015–1019 (2001). https://doi.org/10.1038/nsb725
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb725
This article is cited by
-
1H, 13C, 15N backbone resonance assignment for the 1–164 construct of human XRCC4
Biomolecular NMR Assignments (2021)
-
ATM orchestrates the DNA-damage response to counter toxic non-homologous end-joining at broken replication forks
Nature Communications (2019)
-
Evolutionary diversity and novelty of DNA repair genes in asexual Bdelloid rotifers
BMC Evolutionary Biology (2018)
-
PAXX promotes KU accumulation at DNA breaks and is essential for end-joining in XLF-deficient mice
Nature Communications (2017)
-
Regulation of non-homologous end joining via post-translational modifications of components of the ligation step
Current Genetics (2017)