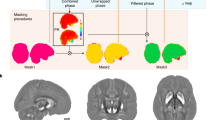
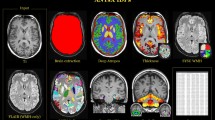
Abstract
Medical imaging has enormous potential for early disease prediction, but is impeded by the difficulty and expense of acquiring data sets before symptom onset. UK Biobank aims to address this problem directly by acquiring high-quality, consistently acquired imaging data from 100,000 predominantly healthy participants, with health outcomes being tracked over the coming decades. The brain imaging includes structural, diffusion and functional modalities. Along with body and cardiac imaging, genetics, lifestyle measures, biological phenotyping and health records, this imaging is expected to enable discovery of imaging markers of a broad range of diseases at their earliest stages, as well as provide unique insight into disease mechanisms. We describe UK Biobank brain imaging and present results derived from the first 5,000 participants' data release. Although this covers just 5% of the ultimate cohort, it has already yielded a rich range of associations between brain imaging and other measures collected by UK Biobank.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Filippini, N. et al. Distinct patterns of brain activity in young carriers of the APOE-epsilon4 allele. Proc. Natl. Acad. Sci. USA 106, 7209–7214 (2009).
Douaud, G. et al. Brain microstructure reveals early abnormalities more than two years prior to clinical progression from mild cognitive impairment to Alzheimer's disease. J. Neurosci. 33, 2147–2155 (2013).
Sudlow, C. et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015).
Allen, N. et al. UK Biobank: current status and what it means for epidemiology. Health Policy and Technology 1, 123–126 (2012).
Wick, J.Y. Understanding frailty in the geriatric population. Consult Pharm. 26, 634–645 (2011).
Stanek, K.M. et al. Obesity is associated with reduced white matter integrity in otherwise healthy adults. Obesity (Silver Spring) 19, 500–504 (2011).
Petersen, S.E. et al. Imaging in population science: cardiovascular magnetic resonance in 100,000 participants of UK Biobank - rationale, challenges and approaches. J. Cardiovasc. Magn. Reson. 15, 46 (2013).
Van Essen, D.C. et al. The WU-Minn Human Connectome Project: an overview. Neuroimage 80, 62–79 (2013).
Good, C.D. et al. A voxel-based morphometric study of ageing in 465 normal adult human brains. Neuroimage 14, 21–36 (2001).
Karas, G.B. et al. A comprehensive study of gray matter loss in patients with Alzheimer's disease using optimized voxel-based morphometry. Neuroimage 18, 895–907 (2003).
Debette, S. & Markus, H.S. The clinical importance of white matter hyperintensities on brain magnetic resonance imaging: systematic review and meta-analysis. Br. Med. J. 341, c3666 (2010).
Haacke, E.M., Xu, Y., Cheng, Y.-C.N. & Reichenbach, J.R. Susceptibility weighted imaging (SWI). Magn. Reson. Med. 52, 612–618 (2004).
Duyn, J. MR susceptibility imaging. J. Magn. Reson. 229, 198–207 (2013).
Beaulieu, C. The basis of anisotropic water diffusion in the nervous system - a technical review. NMR Biomed. 15, 435–455 (2002).
Basser, P.J., Mattiello, J. & LeBihan, D. Estimation of the effective self-diffusion tensor from the NMR spin echo. J. Magn. Reson. B. 103, 247–254 (1994).
Zhang, H., Schneider, T., Wheeler-Kingshott, C.A. & Alexander, D.C. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage 61, 1000–1016 (2012).
Jbabdi, S., Sotiropoulos, S.N., Savio, A.M., Graña, M. & Behrens, T.E. Model-based analysis of multishell diffusion MR data for tractography: how to get over fitting problems. Magn. Reson. Med. 68, 1846–1855 (2012).
de Groot, M. et al. Improving alignment in Tract-based spatial statistics: evaluation and optimization of image registration. Neuroimage 76, 400–411 (2013).
Wakana, S. et al. Reproducibility of quantitative tractography methods applied to cerebral white matter. Neuroimage 36, 630–644 (2007).
Logothetis, N.K. What we can do and what we cannot do with fMRI. Nature 453, 869–878 (2008).
Hariri, A.R., Tessitore, A., Mattay, V.S., Fera, F. & Weinberger, D.R. The amygdala response to emotional stimuli: a comparison of faces and scenes. Neuroimage 17, 317–323 (2002).
Smith, S.M. et al. Functional connectomics from resting-state fMRI. Trends Cogn. Sci. 17, 666–682 (2013).
Douaud, G. et al. DTI measures in crossing-fibre areas: increased diffusion anisotropy reveals early white matter alteration in MCI and mild Alzheimer's disease. Neuroimage 55, 880–890 (2011).
Ennis, D.B. & Kindlmann, G. Orthogonal tensor invariants and the analysis of diffusion tensor magnetic resonance images. Magn. Reson. Med. 55, 136–146 (2006).
Raichle, M.E. et al. A default mode of brain function. Proc. Natl. Acad. Sci. USA 98, 676–682 (2001).
Satterthwaite, T.D. et al. An improved framework for confound regression and filtering for control of motion artifact in the preprocessing of resting-state functional connectivity data. Neuroimage 64, 240–256 (2013).
Kuznetsova, K. et al. Brain white matter structure and information processing speed in healthy older age. Brain Struct. Funct. 221, 3223–3235 (2016).
Madden, D.J. et al. Diffusion tensor imaging of cerebral white matter integrity in cognitive aging. Biochim. Biophys. Acta 1822, 386–400 (2012).
Van Der Werf, Y.D. et al. Thalamic volume predicts performance on tests of cognitive speed and decreases in healthy aging. A magnetic resonance imaging-based volumetric analysis. Brain Res. Cogn. Brain Res. 11, 377–385 (2001).
Fjell, A.M. & Walhovd, K.B. Structural brain changes in aging: courses, causes and cognitive consequences. Rev. Neurosci. 21, 187–221 (2010).
Reichle, E.D., Carpenter, P.A. & Just, M.A. The neural bases of strategy and skill in sentence-picture verification. Cognit. Psychol. 40, 261–295 (2000).
Hotelling, H. Relations between two sets of variates. Biometrika 28, 321–377 (1936).
Hyvärinen, A. Fast and robust fixed-point algorithms for independent component analysis. IEEE Trans. Neural Netw. 10, 626–634 (1999).
Smith, S.M. et al. A positive-negative mode of population covariation links brain connectivity, demographics and behavior. Nat. Neurosci. 18, 1565–1567 (2015).
Smith, S.M. et al. Correspondence of the brain's functional architecture during activation and rest. Proc. Natl. Acad. Sci. USA 106, 13040–13045 (2009).
Yaffe, K., Browner, W., Cauley, J., Launer, L. & Harris, T. Association between bone mineral density and cognitive decline in older women. J. Am. Geriatr. Soc. 47, 1176–1182 (1999).
Tan, Z.S. et al. Bone mineral density and the risk of Alzheimer disease. Arch. Neurol. 62, 107–111 (2005).
Pirpamer, L. et al. Determinants of iron accumulation in the normal aging brain. Neurobiol. Aging 43, 149–155 (2016).
Hamidi, M., Drevets, W.C. & Price, J.L. Glial reduction in amygdala in major depressive disorder is due to oligodendrocytes. Biol. Psychiatry 55, 563–569 (2004).
D'Esposito, M., Deouell, L.Y. & Gazzaley, A. Alterations in the BOLD fMRI signal with ageing and disease: a challenge for neuroimaging. Nat. Rev. Neurosci. 4, 863–872 (2003).
Kirk, R. Practical significance: a concept whose time has come. Educ. Psychol. Meas. 56, 746–759 (1996).
Gage, S.H., Davey Smith, G., Ware, J.J., Flint, J. & Munafò, M.R. G = E: what GWAS can tell us about the environment. PLoS Genet. 12, e1005765 (2016).
Simpson, E. The interpretation of interaction in contingency tables. J. R. Stat. Soc. B 13, 238–241 (1951).
Swanson, J.M. The UK Biobank and selection bias. Lancet 380, 110 (2012).
Berkson, J. Limitations of the application of fourfold table analysis to hospital data. Biometrics 2, 47–53 (1946).
Pearl, J. Causality: Models, Reasoning and Inference (Cambridge University Press, 2009).
Duff, E.P. et al. Learning to identify CNS drug action and efficacy using multistudy fMRI data. Sci. Transl. Med. 7, 274ra16 (2015).
Insel, T. et al. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. Am. J. Psychiatry 167, 748–751 (2010).
Schram, M.T. et al. The Maastricht Study: an extensive phenotyping study on determinants of type 2 diabetes, its complications and its comorbidities. Eur. J. Epidemiol. 29, 439–451 (2014).
German National Cohort (GNC) Consortium. The German National Cohort: aims, study design and organization. Eur. J. Epidemiol. 29, 371–382 (2014).
Jenkinson, M., Beckmann, C.F., Behrens, T.E., Woolrich, M.W. & Smith, S.M. FSL. Neuroimage 62, 782–790 (2012).
Daducci, A. et al. Accelerated Microstructure Imaging via Convex Optimization (AMICO) from diffusion MRI data. Neuroimage 105, 32–44 (2015).
Glasser, M.F. et al. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage 80, 105–124 (2013).
Mennes, M. et al. Optimizing full-brain coverage in human brain MRI through population distributions of brain size. Neuroimage 98, 513–520 (2014).
Andersson, J.L., Skare, S. & Ashburner, J. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. Neuroimage 20, 870–888 (2003).
Uǧurbil, K. et al. Pushing spatial and temporal resolution for functional and diffusion MRI in the Human Connectome Project. Neuroimage 80, 80–104 (2013).
Larkman, D.J. et al. Use of multicoil arrays for separation of signal from multiple slices simultaneously excited. J. Magn. Reson. Imaging 13, 313–317 (2001).
Moeller, S. et al. Multiband multislice GE-EPI at 7 tesla, with 16-fold acceleration using partial parallel imaging with application to high spatial and temporal whole-brain fMRI. Magn. Reson. Med. 63, 1144–1153 (2010).
Setsompop, K. et al. Blipped-controlled aliasing in parallel imaging for simultaneous multislice echo planar imaging with reduced g-factor penalty. Magn. Reson. Med. 67, 1210–1224 (2012).
Feinberg, D. et al. Multiplexed echo planar imaging for sub-second whole brain FMRI and fast diffusion imaging. PLoS One 5, e15710 (2010).
Jenkinson, M., Bannister, P., Brady, M. & Smith, S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 17, 825–841 (2002).
Andersson, J., Jenkinson, M. & Smith, S. Non-linear registration aka spatial normalization. in FMRIB Technical Report (Oxford University, 2007).
Zhang, Y., Brady, M. & Smith, S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans. Med. Imaging 20, 45–57 (2001).
Smith, S.M. et al. Longitudinal and cross-sectional analysis of atrophy in Alzheimer's disease: cross-validation of BSI, SIENA and SIENAX. Neuroimage 36, 1200–1206 (2007).
Patenaude, B., Smith, S.M., Kennedy, D.N. & Jenkinson, M. A Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage 56, 907–922 (2011).
Mugler, J.P. III. Optimized three-dimensional fast-spin-echo MRI. J. Magn. Reson. Imaging 39, 745–767 (2014).
Andersson, J.L. & Sotiropoulos, S.N. Non-parametric representation and prediction of single- and multi-shell diffusion-weighted MRI data using Gaussian processes. Neuroimage 122, 166–176 (2015).
Andersson, J.L. & Sotiropoulos, S.N. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. Neuroimage 125, 1063–1078 (2016).
Hernández, M. et al. Accelerating fibre orientation estimation from diffusion weighted magnetic resonance imaging using GPUs. PLoS One 8, e61892 (2013).
Smith, S.M. et al. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. Neuroimage 31, 1487–1505 (2006).
Wakana, S., Jiang, H., Nagae-Poetscher, L.M., van Zijl, P.C.M. & Mori, S. Fiber tract-based atlas of human white matter anatomy. Radiology 230, 77–87 (2004).
Bannister, P.R., Brady, J.M. & Jenkinson, M. Integrating temporal information with a non-rigid method of motion correction for functional magnetic resonance images. Image Vis. Comput. 25, 311–320 (2007).
Barch, D.M. et al. Function in the human connectome: task-fMRI and individual differences in behavior. Neuroimage 80, 169–189 (2013).
Woolrich, M.W., Ripley, B.D., Brady, M. & Smith, S.M. Temporal autocorrelation in univariate linear modeling of FMRI data. Neuroimage 14, 1370–1386 (2001).
Beckmann, C.F. & Smith, S.M. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans. Med. Imaging 23, 137–152 (2004).
Salimi-Khorshidi, G. et al. Automatic denoising of functional MRI data: combining independent component analysis and hierarchical fusion of classifiers. Neuroimage 90, 449–468 (2014).
Smith, S.M., Hyvärinen, A., Varoquaux, G., Miller, K.L. & Beckmann, C.F. Group-PCA for very large fMRI datasets. Neuroimage 101, 738–749 (2014).
Kiviniemi, V., Kantola, J.-H., Jauhiainen, J., Hyvärinen, A. & Tervonen, O. Independent component analysis of nondeterministic fMRI signal sources. Neuroimage 19, 253–260 (2003).
Kiviniemi, V. et al. Functional segmentation of the brain cortex using high model order group PICA. Hum. Brain Mapp. 30, 3865–3886 (2009).
Smith, S.M. The future of FMRI connectivity. Neuroimage 62, 1257–1266 (2012).
Smith, S.M. et al. Network modelling methods for FMRI. Neuroimage 54, 875–891 (2011).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995).
Genovese, C.R., Lazar, N.A. & Nichols, T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage 15, 870–878 (2002).
Sui, J. et al. A CCA+ICA based model for multi-task brain imaging data fusion and its application to schizophrenia. Neuroimage 51, 123–134 (2010).
Acknowledgements
We would like to acknowledge the valuable contributions of members of the UK Biobank Imaging Working Group and the UK Biobank coordinating center. We are very grateful for additional input into the imaging protocol and image processing pipelines from M. Chappell, S. Clare, E. Duff, D. Flitney, M. Hernandez Fernandez, H. Johansen-Berg, P. McCarthy, J. Miller, D. Mortimer, J. Price, G. Salimi-Khorshidi, E. Vallee, D. Vidaurre, M. Webster, A. Winkler, A. Young, E. Auerbach, S. Moeller, K. Ugurbil, D. Alexander, N. Fox, E. Kaden, S. Ourselin, G. Zhang, A. Daducci, T. Stoecker, D. Barch, N. Bloom, G. Burgess, M. Glasser, M. Harms, D. Nolan, B. Fischl, D. Greve, J. Polimeni, T. Nichols, A. Murphy, G. Parker, F. Barkhof, C. Beckmann, M. Mennes, M. Vernooij, N. Weiskopf, C. Rorden and J. Wardlaw. We are grateful for the provision of simultaneous multi-slice (multiband) pulse sequence and reconstruction algorithms from the Center for Magnetic Resonance Research, University of Minnesota. Finally, we are extremely grateful to all UK Biobank study participants, who generously donated their time to make this resource possible. UK Biobank (including the imaging enhancement) has been generously supported by the UK Medical Research Council and the Wellcome Trust. K.L.M. and S.M.S. receive further support from the Wellcome Trust. P.M.M. acknowledges support from the Edmund J Safra Foundation and Lily Safra, the Imperial College Healthcare Trust Biomedical Research Centre, and the Medical Research Council.
Author information
Authors and Affiliations
Contributions
K.L.M., R.C., P.M.M. and S.M.S. provided the overall scientific strategy for UK Biobank brain imaging. K.L.M., N.K.B., D.L.T., E.Y., J.X., A.J.B., S.J., S.N.S., J.L.R.A., M.J., P.M.M. and S.M.S. developed acquisition protocols. N.K.B., K.L.M., T.W.O., P.W., I.D., S.G. and S.H. implemented the imaging protocol at the dedicated imaging center. F.A.-A., K.L.M., S.J., S.N.S., J.L.R.A., L.G., G.D., M.J. and S.M.S. developed post-processing pipelines and IDP calculation. K.L.M. and S.M.S. carried out the univariate and multivariate analyses and prepared figures. K.L.M. and S.M.S. wrote the manuscript, which was edited by all of the authors.
Corresponding author
Ethics declarations
Competing interests
P.W. and I.D. are employees of Siemens Healthcare UK, the vendor of MRI scanners for UK Biobank, selected under a competitive bidding process.
Integrated supplementary information
Supplementary Figure 1 Asymptotic behavior of group average images and correlations with age, with increasing numbers of subjects.
(a) Resting-state network group-averages formed from 5 different group sizes. Individual subjects' preprocessed resting-state data were used to generate subject-specific effect-size maps (arbitrary but consistent units of resting activity strength) of one of the low-dimensional resting-state networks (the default mode network, here shown with both positive=red and negative=blue involvement in this network, with the same color-coding and thresholding applied in all cases). These were then averaged across subjects for a range of subject numbers. Increasing n suppresses background noise as expected, and the non-noise network structure asymptotes towards a constant map as n rises over 100. Although any imperfections in functional spatial alignment across subjects limits the sharpness of the asymptotic group-average, as n is raised even further to 5000, there is no sign of a degradation (e.g., blurring) of the group-average map (compared with lower subject numbers), as expected. (b-d) Voxelwise correlation of the same resting-state network with increasing subject age, again to illustrate the statistical effect of increasing the number of subjects used. (b) The age correlation map when using 5000 subjects; with increasing age the correlations are dominantly negative, indicating a weakening of this cognitive network (r>0.1, Pcorrected<10-10). (c) The 10 voxels having the strongest (positive or negative) correlation with age, that are also at least 10mm distant from each other, are used to form 10 plots of age-correlation against number of subjects used in the correlation, from 10 to 5000 subjects. As expected, the plots asymptote, with increasing subject numbers, towards the "true" final value, with noticeable instability up to as many as 2000 subjects. (d) From the same 10 sets of correlations, the statistical significance (-log10(P)) is shown. Whereas r asymptotes towards its true final constant value with increasing n, statistical significance (for a non-null correlation) has an ever-increasing trend with increasing n. (e) Theoretical relationship between increasing statistical power and subject numbers, assuming a true correlation between any two variables of r=0.1. As n increases (here up to 7000), the number of distinct tests that will pass Bonferroni multiple-comparison correction rises to very large numbers - here up to around 1015.
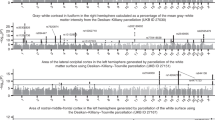
Supplementary Figure 2 Visualisation of 2.8 million univariate cross-subject association tests between 2501 IDPs and 1100 other variables in the UK Biobank database, showing variance explained on the y axis.
Whereas the version of this plot in Figure 6 reported statistical significance (-log10(P)), here we show variance explained (r2). The relationship between P and r is not here exactly a fixed one-to-one mapping, due to the different numbers of valid (non-missing) data in different pairs of variables being tested. The Manhattan plot shows, for each of the 1100 non-brain-imaging variables, the strongest r2 association of that variable with each distinct imaging sub-modality’s IDPs. (i.e., 6 results plotted for each x axis position, each with a color indicating a brain imaging modality; this plot differs from the other Manhattan plots, which show correlations with all IDPs).
Supplementary Figure 3 Visualisation of modes 1, 2 and 3 from the doubly-multivariate CCA-ICA analyses across all IDPs and non-brain-imaging variables.
(a) Mode 1 links physical measures of body size, metabolic rate and hand grip strength to brain structure sizes and a range of dMRI-derived measures. (b) Mode 2 primarily links bone density measures to brain structure sizes, T2* levels and a range of dMRI-derived measures. (c) Mode 3 primarily links measures of body fat to T2* levels and resting-state activity fluctuation amplitudes. As seen in Supp Fig 8a, modes 1 and 2 are associated with aging (and sex), while mode 3 is not strongly associated; from Supp Fig 8b, we see that modes 1 and (more strongly) 3 are associated with hypertension.
Supplementary Figure 4 Visualisation of modes 4, 5 and 6 from the doubly-multivariate CCA-ICA analyses across all IDPs and non-brain-imaging variables.
(a) Mode 4 links cardiac measures (including heart rate) to resting-state amplitudes and connectivities (rfMRI summary images are shown larger in Supp Fig 5). Observing these 3 types of measures linked together suggests that the apparent change in functional connectivity in this mode likely reflects changes in vascular processes rather than underlying neural connectivity. (b) Mode 5 links a range of lifestyle and biophysical measures (most strongly alcohol intake and smoking, red blood cell and cardiac measures) to T2* subcortical intensity (e.g., iron deposition) and the resting-state amplitudes of many brain areas (rfMRI summary images shown larger in Supp Fig 6). (c) Mode 6 links early-life measures (birth weight and breast feeding) along with several other physical and lifestyle measures to many imaging measures of both diffusivity and functional connectivity (rfMRI summary images are shown larger in Supp Fig 7).
Supplementary Figure 8 Associations of the nine CCA-ICA modes with confounds and other variables of interest.
(a) As with the univariate analyses, data fed into the multivariate analyses were first adjusted for parameters that might otherwise induce apparent relationships based on potentially non-interesting factors (age, sex, head size, head motion). Here we show how, by projecting the CCA-ICA modes back onto the original data, we can estimate how strongly the discovered modes relate to these parameters. For example, modes 1,2,4,5,7,8 are associated with aging, whereas 3,6,9 are not strongly associated with age. Considering this in the light of the fact that all data input to the CCA had been age-adjusted suggests that, while these modes reflect meaningful biological processes related to aging, their identification here was not driven by trivial corruption of IDPs by aging (e.g., reduced fMRI signal due to cortical thinning). (b) Correlation of the CCA-ICA modes against several variables of interest, including some clinical outcomes (for which, at this stage, numbers are naturally quite limited). (c) Scatterplot of CCA-ICA modes 1 vs. 8 (one point per subject), showing the associations between these modes, age and sex. Colors running from green to red indicate increasing age in females; colors running from blue to pink indicate increasing age in males.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Table 1 (PDF 3206 kb)
Rights and permissions
About this article
Cite this article
Miller, K., Alfaro-Almagro, F., Bangerter, N. et al. Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat Neurosci 19, 1523–1536 (2016). https://doi.org/10.1038/nn.4393
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.4393