Abstract
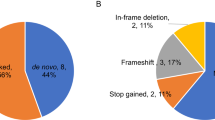
Evidence for the etiology of autism spectrum disorders (ASDs) has consistently pointed to a strong genetic component complicated by substantial locus heterogeneity1,2. We sequenced the exomes of 20 individuals with sporadic ASD (cases) and their parents, reasoning that these families would be enriched for de novo mutations of major effect. We identified 21 de novo mutations, 11 of which were protein altering. Protein-altering mutations were significantly enriched for changes at highly conserved residues. We identified potentially causative de novo events in 4 out of 20 probands, particularly among more severely affected individuals, in FOXP1, GRIN2B, SCN1A and LAMC3. In the FOXP1 mutation carrier, we also observed a rare inherited CNTNAP2 missense variant, and we provide functional support for a multi-hit model for disease risk3. Our results show that trio-based exome sequencing is a powerful approach for identifying new candidate genes for ASDs and suggest that de novo mutations may contribute substantially to the genetic etiology of ASDs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Change history
09 March 2012
In the version of this article initially published, the missense variant in CNTNAP2 identified in proband 12817 was incorrectly listed as p.His275Ala in the main text and Figure 1. The correct notation for this variant is p.His275Arg. In addition, the raw sequence reads have now been deposited in the National Database for Autism Research under accession number NDARCOL0001878. These errors have been corrected in the HTML and PDF versions of the article.
References
Bailey, A. et al. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol. Med. 25, 63–77 (1995).
O'Roak, B.J. & State, M.W. Autism genetics: strategies, challenges, and opportunities. Autism Res. 1, 4–17 (2008).
Girirajan, S. et al. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat. Genet. 42, 203–209 (2010).
Abrahams, B.S. & Geschwind, D.H. Advances in autism genetics: on the threshold of a new neurobiology. Nat. Rev. Genet. 9, 341–355 (2008).
Sebat, J. et al. Strong association of de novo copy number mutations with autism. Science 316, 445–449 (2007).
Marshall, C.R. et al. Structural variation of chromosomes in autism spectrum disorder. Am. J. Hum. Genet. 82, 477–488 (2008).
Durkin, M.S. et al. Advanced parental age and the risk of autism spectrum disorder. Am. J. Epidemiol. 168, 1268–1276 (2008).
Ng, S.B. et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461, 272–276 (2009).
Bailey, J.A. et al. Recent segmental duplications in the human genome. Science 297, 1003–1007 (2002).
de Kovel, C.G. et al. Recurrent microdeletions at 15q11.2 and 16p13.11 predispose to idiopathic generalized epilepsies. Brain 133, 23–32 (2010).
Stefansson, H. et al. Large recurrent microdeletions associated with schizophrenia. Nature 455, 232–236 (2008).
Kirov, G. et al. Support for the involvement of large CNVs in the pathogenesis of schizophrenia. Hum. Mol. Genet. 18, 1497–1503 (2009).
Vissers, L.E. et al. A de novo paradigm for mental retardation. Nat. Genet. 42, 1109–1112 (2010).
Lynch, M. Rate, molecular spectrum, and consequences of human mutation. Proc. Natl. Acad. Sci. USA 107, 961–968 (2010).
Durbin, R.M. et al. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
Adzhubei, I.A. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010).
Grantham, R. Amino acid difference formula to help explain protein evolution. Science 185, 862–864 (1974).
Cooper, G.M. et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 15, 901–913 (2005).
Cooper, G.M. et al. Single-nucleotide evolutionary constraint scores highlight disease-causing mutations. Nat. Methods 7, 250–251 (2010).
Gotham, K., Pickles, A. & Lord, C. Standardizing ADOS scores for a measure of severity in autism spectrum disorders. J. Autism Dev. Disord. 39, 693–705 (2009).
Endele, S. et al. Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat. Genet. 42, 1021–1026 (2010).
Claes, L. et al. De novo mutations in the sodium-channel gene SCN1A cause severe myoclonic epilepsy of infancy. Am. J. Hum. Genet. 68, 1327–1332 (2001).
Weiss, L.A. et al. Sodium channels SCN1A, SCN2A and SCN3A in familial autism. Mol. Psychiatry 8, 186–194 (2003).
Mulley, J.C. et al. SCN1A mutations and epilepsy. Hum. Mutat. 25, 535–542 (2005).
Lein, E.S. et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007).
Hamdan, F.F. et al. De novo mutations in FOXP1 in cases with intellectual disability, autism, and language impairment. Am. J. Hum. Genet. 87, 671–678 (2010).
Horn, D. et al. Identification of FOXP1 deletions in three unrelated patients with mental retardation and significant speech and language deficits. Hum. Mutat. 31, E1851–E1860 (2010).
Lai, C.S., Fisher, S.E., Hurst, J.A., Vargha-Khadem, F. & Monaco, A.P. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature 413, 519–523 (2001).
Feuk, L. et al. Absence of a paternally inherited FOXP2 gene in developmental verbal dyspraxia. Am. J. Hum. Genet. 79, 965–972 (2006).
MacDermot, K.D. et al. Identification of FOXP2 truncation as a novel cause of developmental speech and language deficits. Am. J. Hum. Genet. 76, 1074–1080 (2005).
Vernes, S.C. et al. Functional genetic analysis of mutations implicated in a human speech and language disorder. Hum. Mol. Genet. 15, 3154–3167 (2006).
Li, S., Weidenfeld, J. & Morrisey, E.E. Transcriptional and DNA binding activity of the Foxp1/2/4 family is modulated by heterotypic and homotypic protein interactions. Mol. Cell. Biol. 24, 809–822 (2004).
Teramitsu, I., Kudo, L.C., London, S.E., Geschwind, D.H. & White, S.A. Parallel FoxP1 and FoxP2 expression in songbird and human brain predicts functional interaction. J. Neurosci. 24, 3152–3163 (2004).
Bakkaloglu, B. et al. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am. J. Hum. Genet. 82, 165–173 (2008).
Vernes, S.C. et al. A functional genetic link between distinct developmental language disorders. N. Engl. J. Med. 359, 2337–2345 (2008).
Arking, D.E. et al. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am. J. Hum. Genet. 82, 160–164 (2008).
Alarcón, M. et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am. J. Hum. Genet. 82, 150–159 (2008).
Banerjee-Basu, S. & Packer, A. SFARI Gene: an evolving database for the autism research community. Dis. Model Mech. 3, 133–135 (2010).
Fischbach, G.D. & Lord, C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron 68, 192–195 (2010).
Hurley, R.S., Losh, M., Parlier, M., Reznick, J.S. & Piven, J. The broad autism phenotype questionnaire. J. Autism Dev. Disord. 37, 1679–1690 (2007).
Constantino, J.N. & Todd, R.D. Intergenerational transmission of subthreshold autistic traits in the general population. Biol. Psychiatry 57, 655–660 (2005).
Selzer, R.R. et al. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosom. Cancer 44, 305–319 (2005).
Itsara, A. et al. Population analysis of large copy number variants and hotspots of human genetic disease. Am. J. Hum. Genet. 84, 148–161 (2009).
Igartua, C. et al. Targeted enrichment of specific regions in the human genome by array hybridization. Curr. Protoc. Hum. Genet. Chapter 18, Unit 18 3 (2010).
Ng, S.B. et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 42, 790–793 (2010).
Roach, J.C. et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science 328, 636–639 (2010).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Bailey, J.A., Yavor, A.M., Massa, H.F., Trask, B.J. & Eichler, E.E. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 11, 1005–1017 (2001).
Sudmant, P.H. et al. Diversity of human copy number variation and multicopy genes. Science 330, 641–646 (2010).
Hach, F. et al. mrsFAST: a cache-oblivious algorithm for short-read mapping. Nat. Methods 7, 576–577 (2010).
Andrés, A.M. et al. Balancing selection maintains a form of ERAP2 that undergoes nonsense-mediated decay and affects antigen presentation. PLoS Genet. 6, e1001157 (2010).
Acknowledgements
We would like to thank and recognize the following ongoing studies that produced and provided exome variant calls for comparison: National Heart, Lung, and Blood Institute (NHBLI) Lung Cohort Sequencing Project (HL 1029230), NHLBI Women's Health Initiative (WHI) Sequencing Project (HL 102924), National Institute of Environmental Health Sciences (NIEHS) SNPs (HHSN273200800010C), NHLBI/National Human Genome Research Institute (NHGRI) SeattleSeq (HL 094976), NHGRI Next Generation Mendelian Genetics (HG 005608) and the Northwest Genomics Center (HL 102926). We also thank M.-C. King and S. Stray for processing and managing DNA samples, B.H. King and E. Bliss for their work in subject recruitment and phenotype collection, E. Turner, C. Igartua, I. Stanaway, M. Dennis and B. Coe for thoughtful discussions, M. State for providing SNP genotyping data and especially the families that volunteered their time to participate in this research. This work was supported by US National Institutes of Health grant HD065285 (E.E.E. and J.S.), Wellcome Trust core award 075491/Z/04 (S.E.F. and P.D.), the Max Planck Society (S.E.F.) and grants from the Simons Foundation Autism Research Initiative (SFARI) (191889, 137578 and 137593) (E.E.E., R.B., S.E.F. and P.D.). E.E.E. is an Investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
E.E.E., J.S. and B.J.O. designed the study and drafted the manuscript. E.E.E. and J.S. supervised the study. R.B. analyzed the clinical information and contributed to the manuscript. S.E.F. and P.D. designed cell-based functional experiments, analyzed data, interpreted results and contributed to the manuscript. S.G., C.B. and L.V. generated and analyzed array CGH data. C.L. performed Illumina GAIIx sequencing. B.J.O. and E.K. developed the analysis pipeline and analyzed sequence data. A.P.M. and S.B.N. designed and optimized capture protocol. B.J.O., L.V., A.P.M. and S.B.N. constructed exome libraries. B.J.O., L.V., A.P.M. and J.J.S. performed mutation validation and haplotype characterization. B.J.O. and J.J.S. performed the evaluation of 12817 lymphoblast cell lines. P.D. performed functional experiments. M.J.R. and D.A.N. performed sequencing of control samples.
Corresponding authors
Ethics declarations
Competing interests
E.E.E. is on the scientific advisory board for Pacific Biosciences. J.S. is a member of the scientific advisory boards of Tandem Technologies, Stratos Genomics, Good Start Genetics, Halo Genomics and Adaptive TCR. B.J.O. is an inventor on patent PCT/US2009/30620: Mutations in Contactin Associated Protein 2 are Associated with Increased Risk for Idiopathic Autism.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Figures 1–5 and Supplementary Tables 2–4 and 6–9. (PDF 5445 kb)
Supplementary Table 1
Core descriptive clinical values on ASD probands (XLSX 34 kb)
Supplementary Table 5
Variant positions of 21 genes with identified de novo events from 1000 genomes pilot data, 20 HapMap, and 20 ASD probands (XLSX 126 kb)
Rights and permissions
About this article
Cite this article
O'Roak, B., Deriziotis, P., Lee, C. et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet 43, 585–589 (2011). https://doi.org/10.1038/ng.835
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.835
This article is cited by
-
Possible roles of deep cortical neurons and oligodendrocytes in the neural basis of human sociality
Anatomical Science International (2024)
-
Non-synaptic function of the autism spectrum disorder-associated gene SYNGAP1 in cortical neurogenesis
Nature Neuroscience (2023)
-
Overexpression of EphB6 and EphrinB2 controls soma spacing of cortical neurons in a mutual inhibitory way
Cell Death & Disease (2023)
-
A novel autism-associated UBLCP1 mutation impacts proteasome regulation/activity
Translational Psychiatry (2023)
-
Inhibition of Foxp4 Disrupts Cadherin-based Adhesion of Radial Glial Cells, Leading to Abnormal Differentiation and Migration of Cortical Neurons in Mice
Neuroscience Bulletin (2023)