Abstract
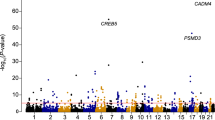
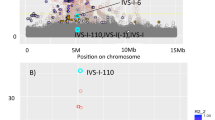
Hematologic measures such as hematocrit and white blood cell (WBC) count are heritable and clinically relevant. We analyzed erythrocyte and WBC phenotypes in 52,531 individuals (37,775 of European ancestry, 11,589 African Americans, and 3,167 Hispanic Americans) from 16 population-based cohorts with Illumina HumanExome BeadChip genotypes. We then performed replication analyses of new discoveries in 18,018 European-American women and 5,261 Han Chinese. We identified and replicated four new erythrocyte trait–locus associations (CEP89, SHROOM3, FADS2, and APOE) and six new WBC loci for neutrophil count (S1PR4), monocyte count (BTBD8, NLRP12, and IL17RA), eosinophil count (IRF1), and total WBC count (MYB). The association of a rare missense variant in S1PR4 supports the role of sphingosine-1-phosphate signaling in leukocyte trafficking and circulating neutrophil counts. Loss-of-function experiments for S1pr4 in mouse and s1pr4 in zebrafish demonstrated phenotypes consistent with the association observed in humans and altered kinetics of neutrophil recruitment and resolution in response to tissue injury.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Whitfield, J.B. & Martin, N.G. Genetic and environmental influences on the size and number of cells in the blood. Genet. Epidemiol. 2, 133–144 (1985).
Evans, D.M., Frazer, I.H. & Martin, N.G. Genetic and environmental causes of variation in basal levels of blood cells. Twin Res. 2, 250–257 (1999).
Lin, J.P. et al. Evidence for linkage of red blood cell size and count: genome-wide scans in the Framingham Heart Study. Am. J. Hematol. 82, 605–610 (2007).
Pilia, G. et al. Heritability of cardiovascular and personality traits in 6,148 Sardinians. PLoS Genet. 2, e132 (2006).
Zakai, N.A. et al. A prospective study of anemia status, hemoglobin concentration, and mortality in an elderly cohort: the Cardiovascular Health Study. Arch. Intern. Med. 165, 2214–2220 (2005).
Brennan, M.L. et al. Comprehensive peroxidase-based hematologic profiling for the prediction of 1-year myocardial infarction and death. Circulation 122, 70–79 (2010).
Elwood, P.C., Waters, W.E., Benjamin, I.T. & Sweetnam, P.M. Mortality and anaemia in women. Lancet 1, 891–894 (1974).
Reiner, A.P. et al. Genome-wide association study of white blood cell count in 16,388 African Americans: the continental origins and genetic epidemiology network (COGENT). PLoS Genet. 7, e1002108 (2011).
Ganesh, S.K. et al. Multiple loci influence erythrocyte phenotypes in the CHARGE Consortium. Nat. Genet. 41, 1191–1198 (2009).
van der Harst, P. et al. Seventy-five genetic loci influencing the human red blood cell. Nature 492, 369–375 (2012).
Kamatani, Y. et al. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat. Genet. 42, 210–215 (2010).
Pistis, G. et al. Genome wide association analysis of a founder population identified TAF3 as a gene for MCHC in humans. PLoS One 8, e69206 (2013).
Nalls, M.A. et al. Multiple loci are associated with white blood cell phenotypes. PLoS Genet. 7, e1002113 (2011).
Okada, Y. et al. Identification of nine novel loci associated with white blood cell subtypes in a Japanese population. PLoS Genet. 7, e1002067 (2011).
Auer, P.L. et al. Imputation of exome sequence variants into population- based samples and blood-cell-trait-associated loci in African Americans: NHLBI GO Exome Sequencing Project. Am. J. Hum. Genet. 91, 794–808 (2012).
Auer, P.L. et al. Rare and low-frequency coding variants in CXCR2 and other genes are associated with hematological traits. Nat. Genet. 46, 629–634 (2014).
Okada, Y. et al. Meta-analysis identifies multiple loci associated with kidney function–related traits in East Asian populations. Nat. Genet. 44, 904–909 (2012).
Meyer, T.E. et al. Genome-wide association studies of serum magnesium, potassium, and sodium concentrations identify six loci influencing serum magnesium levels. PLoS Genet. 6, e1001045 (2010).
Chambers, J.C. et al. Genetic loci influencing kidney function and chronic kidney disease. Nat. Genet. 42, 373–375 (2010).
Benyamin, B. et al. Novel loci affecting iron homeostasis and their effects in individuals at risk for hemochromatosis. Nat. Commun. 5, 4926 (2014).
Lemaitre, R.N. et al. Genetic loci associated with plasma phospholipid n-3 fatty acids: a meta-analysis of genome-wide association studies from the CHARGE Consortium. PLoS Genet. 7, e1002193 (2011).
Wu, Y. et al. Genetic association with lipids in Filipinos: waist circumference modifies an APOA5 effect on triglyceride levels. J. Lipid Res. 54, 3198–3205 (2013).
Rasmussen-Torvik, L.J. et al. High density GWAS for LDL cholesterol in African Americans using electronic medical records reveals a strong protective variant in APOE. Clin. Transl. Sci. 5, 394–399 (2012).
Chasman, D.I. et al. Genetic determinants of statin-induced low-density lipoprotein cholesterol reduction: the Justification for the Use of Statins in Prevention: an Intervention Trial Evaluating Rosuvastatin (JUPITER) trial. Circ Cardiovasc Genet 5, 257–264 (2012).
Kettunen, J. et al. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat. Genet. 44, 269–276 (2012).
Corder, E.H. et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat. Genet. 7, 180–184 (1994).
Talbot, C. et al. Protection against Alzheimer's disease with apoE ɛ2. Lancet 343, 1432–1433 (1994).
Adzhubei, I.A. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010).
Moffatt, M.F. et al. A large-scale, consortium-based genomewide association study of asthma. N. Engl. J. Med. 363, 1211–1221 (2010).
Barrett, J.C. et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat. Genet. 40, 955–962 (2008).
McGovern, D.P. et al. Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn's disease. Hum. Mol. Genet. 19, 3468–3476 (2010).
Jostins, L. et al. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012).
Sabater-Lleal, M. et al. Multiethnic meta-analysis of genome-wide association studies in >100 000 subjects identifies 23 fibrinogen-associated loci but no strong evidence of a causal association between circulating fibrinogen and cardiovascular disease. Circulation 128, 1310–1324 (2013).
Zhang, X. et al. Synthesis of 53 tissue and cell line expression QTL datasets reveals master eQTLs. BMC Genomics 15, 532 (2014).
Allende, M.L. et al. Sphingosine-1-phosphate lyase deficiency produces a pro-inflammatory response while impairing neutrophil trafficking. J. Biol. Chem. 286, 7348–7358 (2011).
Allende, M.L. et al. S1P1 receptor directs the release of immature B cells from bone marrow into blood. J. Exp. Med. 207, 1113–1124 (2010).
Amatruda, J.F. & Zon, L.I. Dissecting hematopoiesis and disease using the zebrafish. Dev. Biol. 216, 1–15 (1999).
Rivera, J., Proia, R.L. & Olivera, A. The alliance of sphingosine-1-phosphate and its receptors in immunity. Nat. Rev. Immunol. 8, 753–763 (2008).
Allende, M.L., Dreier, J.L., Mandala, S. & Proia, R.L. Expression of the sphingosine 1-phosphate receptor, S1P1, on T-cells controls thymic emigration. J. Biol. Chem. 279, 15396–15401 (2004).
Matloubian, M. et al. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature 427, 355–360 (2004).
Schwab, S.R. & Cyster, J.G. Finding a way out: lymphocyte egress from lymphoid organs. Nat. Immunol. 8, 1295–1301 (2007).
Golfier, S. et al. Shaping of terminal megakaryocyte differentiation and proplatelet development by sphingosine-1-phosphate receptor S1P4 . FASEB J. 24, 4701–4710 (2010).
Schulze, T. et al. Sphingosine-1-phospate receptor 4 (S1P) deficiency profoundly affects dendritic cell function and TH17-cell differentiation in a murine model. FASEB J. 25, 4024–4036 (2011).
Dillmann, C., Mora, J., Olesch, C., Brüne, B. & Weigert, A. S1PR4 is required for plasmacytoid dendritic cell differentiation. Biol. Chem. 396, 775–782 (2015).
Olivera, A. et al. Sphingosine kinase 1 and sphingosine-1-phosphate receptor 2 are vital to recovery from anaphylactic shock in mice. J. Clin. Invest. 120, 1429–1440 (2010).
Eash, K.J., Greenbaum, A.M., Gopalan, P.K. & Link, D.C. CXCR2 and CXCR4 antagonistically regulate neutrophil trafficking from murine bone marrow. J. Clin. Invest. 120, 2423–2431 (2010).
Beck, T.C., Gomes, A.C., Cyster, J.G. & Pereira, J.P. CXCR4 and a cell-extrinsic mechanism control immature B lymphocyte egress from bone marrow. J. Exp. Med. 211, 2567–2581 (2014).
McEver, R.P., Moore, K.L. & Cummings, R.D. Leukocyte trafficking mediated by selectin–carbohydrate interactions. J. Biol. Chem. 270, 11025–11028 (1995).
Ye, Z. et al. ATP binding by monarch-1/NLRP12 is critical for its inhibitory function. Mol. Cell. Biol. 28, 1841–1850 (2008).
Borghini, S. et al. Clinical presentation and pathogenesis of cold-induced autoinflammatory disease in a family with recurrence of an NLRP12 mutation. Arthritis Rheum. 63, 830–839 (2011).
Landrum, M.J. et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 42, D980–D985 (2014).
Arthur, J.C. et al. Cutting edge: NLRP12 controls dendritic and myeloid cell migration to affect contact hypersensitivity. J. Immunol. 185, 4515–4519 (2010).
Gaffen, S.L. An overview of IL-17 function and signaling. Cytokine 43, 402–407 (2008).
Butcher, M.J., Gjurich, B.N., Phillips, T. & Galkina, E.V. The IL-17A/IL-17RA axis plays a proatherogenic role via the regulation of aortic myeloid cell recruitment. Circ. Res. 110, 675–687 (2012).
Puel, A. et al. Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science 332, 65–68 (2011).
Hashiguchi, M. et al. IL-33 activates eosinophils of visceral adipose tissue both directly and via innate lymphoid cells. Eur. J. Immunol. 45, 876–885 (2015).
Franke, A. et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat. Genet. 42, 1118–1125 (2010).
Kouro, T. & Takatsu, K. IL-5– and eosinophil-mediated inflammation: from discovery to therapy. Int. Immunol. 21, 1303–1309 (2009).
Tin, A. et al. Using multiple measures for quantitative trait association analyses: application to estimated glomerular filtration rate. J. Hum. Genet. 58, 461–466 (2013).
Li, J. et al. Piezo1 integration of vascular architecture with physiological force. Nature 515, 279–282 (2014).
Ranade, S.S. et al. Piezo1, a mechanically activated ion channel, is required for vascular development in mice. Proc. Natl. Acad. Sci. USA 111, 10347–10352 (2014).
Peyronnet, R. et al. Piezo1-dependent stretch-activated channels are inhibited by Polycystin-2 in renal tubular epithelial cells. EMBO Rep. 14, 1143–1148 (2013).
Miyamoto, T. et al. Functional role for Piezo1 in stretch-evoked Ca2+ influx and ATP release in urothelial cell cultures. J. Biol. Chem. 289, 16565–16575 (2014).
Brohawn, S.G., Su, Z. & MacKinnon, R. Mechanosensitivity is mediated directly by the lipid membrane in TRAAK and TREK1 K+ channels. Proc. Natl. Acad. Sci. USA 111, 3614–3619 (2014).
Sandberg, M.B., Nybo, M., Birgens, H. & Frederiksen, H. Hereditary xerocytosis and familial haemolysis due to mutation in the PIEZO1 gene: a simple diagnostic approach. Int. J. Lab. Hematol. 36, e62–e65 (2014).
Yeo, N.C. et al. Shroom3 contributes to the maintenance of the glomerular filtration barrier integrity. Genome Res. 25, 57–65 (2015).
Feliubadaló, L. et al. Non–type I cystinuria caused by mutations in SLC7A9, encoding a subunit (bo,+AT) of rBAT. Nat. Genet. 23, 52–57 (1999).
Teslovich, T.M. et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 (2010).
van den Berg, J.J. et al. Increased n-3 polyunsaturated fatty acid content of red blood cells from fish oil–fed rabbits increases in vitro lipid peroxidation, but decreases hemolysis. Free Radic. Biol. Med. 11, 393–399 (1991).
Waldron, T. et al. c-Myb and its target Bmi1 are required for p190BCR/ABL leukemogenesis in mouse and human cells. Leukemia 26, 644–653 (2012).
Schunkert, H. et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 43, 333–338 (2011).
Stahl, E.A. et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet. 42, 508–514 (2010).
Levy, D. et al. Genome-wide association study of blood pressure and hypertension. Nat. Genet. 41, 677–687 (2009).
Shameer, K. et al. A genome- and phenome-wide association study to identify genetic variants influencing platelet count and volume and their pleiotropic effects. Hum. Genet. 133, 95–109 (2014).
Plagnol, V. et al. Genome-wide association analysis of autoantibody positivity in type 1 diabetes cases. PLoS Genet. 7, e1002216 (2011).
Dichgans, M. et al. Shared genetic susceptibility to ischemic stroke and coronary artery disease: a genome-wide analysis of common variants. Stroke 45, 24–36 (2014).
Psaty, B.M. et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet 2, 73–80 (2009).
Liu, X., Jian, X. & Boerwinkle, E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum. Mutat. 32, 894–899 (2011).
Liu, X., Jian, X. & Boerwinkle, E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum. Mutat. 34, E2393–E2402 (2013).
Grove, M.L. et al. Best practices and joint calling of the HumanExome BeadChip: the CHARGE Consortium. PLoS One 8, e68095 (2013).
Li, B. & Leal, S.M. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am. J. Hum. Genet. 83, 311–321 (2008).
Wu, M.C. et al. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 89, 82–93 (2011).
Johnson, A.D. et al. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 24, 2938–2939 (2008).
Bain, B.J. & England, J.M. Normal haematological values: sex difference in neutrophil count. BMJ 1, 306–309 (1975).
Bain, B.J. & England, J.M. Variations in leucocyte count during menstrual cycle. BMJ 2, 473–475 (1975).
Acknowledgements
We thank the staff and participants of all studies for their important contributions. A complete list of acknowledgments for each study is available in the Supplementary Note.
This work was supported by the following grants and contracts: US National Institutes of Health contracts (N01AG12100, HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, HHSN268201100012C, HHSN268201200036C, HHSN268200800007C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086, N01HC25195, N02HL64278, N01AG62101, N01AG62103, N01AG62106, HHSN268200782096C, HHSN268201300046C, HHSN268201300047C, HHSN268201300048C, HHSN268201300049C, HHSN268201300050C, N01HC95159, N01HC95160, N01HC95161, N01HC95162, N01HC95163, N01HC95164, N01HC95165, N01HC95166, N01HC95167, N01HC95168, N01HC95169, RR024156, N02HL64278, HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C, HHSN271201100004C, RC2HL102924, and CA137088); US National Institutes of Health grants (5RC2HL102419, HL080295, HL087652, HL103612, HL105756, HL120393, AG023629, DK063491, R01DK089256, R01HL087700, R01HL088215, R01HL117078, 1R01AG032098-01A1, U01-HG005152, R25CA094880, R01HL122684, R01HL04880, R01HL32262, R01DK49216, R01HL10001, R01DK092760, and R01OD017870); a Clinical and Translational Science Institute grant (UL1TR000124); a Danish Heart Foundation grant (07-10-R61-A1754-B838-22392F); a Biobanking and BioMolecular resources Research Infrastructure–The Netherlands (BBMRI-NL) grant (NWO 184.021.007); a Health Insurance Foundation grant (2012B233); and Academy of Finland grants (134309, 126925, 121584, 124282, 129378, 117787, and 41071).
This work was supported in part by the NIDDK Division of Intramural Research.
The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute, the National Institutes of Health, or the US Department of Health and Human Services.
This work was carried out in part using computing resources at the University of Minnesota Supercomputing Institute.
Author information
Authors and Affiliations
Consortia
Contributions
N.P., Y. Zhou, Y. Zhang, E.P.B., I.J.D., O.H.F., M.E.G., V.G., T.H., T.B.H., A.H., L.J.L., A.L., O.P., J.M.S., A.D., Y.H., C.J.O'D., A.P.R., and S.K.G. designed the study. Y. Zhang, I.B.B., E.P.B., M.C., I.J.D., L.D., M.F.F., M.E.G., V.G., T.B.H., A.H., R.D.J., J.J., M.K., T.L., A.L., M.E.N., B.M.P., O.T.R., S.S.R., J.M.S., B.H.T., R.P.T., Jiansong Wang, and C.J.O'D. recruited and assessed participants. P.L.A., J.B.-J., N.G., L.-P.L., Y. Zhang, F.W.A., E.B., I.B.B., E.P.B., P.I.W.d.B., M.F.F., M.L.G., T.L., D.C.L., Y. Liu, S.S.R., F.R., J.I.R., K.D.T., and A.G.U. generated genotyping data. Y. Zhou, M.L.A., V.C., E.J.H., B.H., K.H., X.Z., V.M.N., A.M.R.D.S., R.L.P., and L.I.Z. performed functional experiments. N.P., U.M.S., T.S.A., M.L.A., P.L.A., J.B.-J., N.G., B.H., Y. Lu, M.A.N., R.P., A.V.S., Y. Zhang, J.S.F., N.F., M.L.G., R.J.F.L., B.M.P., A.D., L.A.C., J.G.W., R.L.P., L.I.Z., C.J.O'D., A.P.R., and S.K.G. analyzed and interpreted data. N.P., U.M.S., W.Z., T.S.A., J.B.-J., J.A.B., M.-H.C., J.D.E., N.G., A.D.J., M.L., Y. Lu, L.-P.L., A.M., R.E.M., M.A.N., R.P., A.V.S., F.J.A.v.R., M.-L.Y., Judy Wang, and A.P.R. performed statistical analysis. N.P., U.M.S., Y. Zhou, A.P.R., and S.K.G. wrote the manuscript. All authors were given the opportunity to comment and provide revisions to the manuscript text.
Corresponding authors
Ethics declarations
Competing interests
The author declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Note. (PDF 2138 kb)
Supplementary Tables 1–16
Supplementary Tables 1–16. (XLSX 619 kb)
Rights and permissions
About this article
Cite this article
the CHARGE Consortium Hematology Working Group. Meta-analysis of rare and common exome chip variants identifies S1PR4 and other loci influencing blood cell traits. Nat Genet 48, 867–876 (2016). https://doi.org/10.1038/ng.3607
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3607
This article is cited by
-
Insights into the genetic architecture of haematological traits from deep phenotyping and whole-genome sequencing for two Mediterranean isolated populations
Scientific Reports (2022)
-
Sphingosine 1-phosphate receptor-targeted therapeutics in rheumatic diseases
Nature Reviews Rheumatology (2022)
-
Sphingosine 1-Phosphate Receptors in Cerebral Ischemia
NeuroMolecular Medicine (2021)
-
Common variants in signaling transcription-factor-binding sites drive phenotypic variability in red blood cell traits
Nature Genetics (2020)
-
Genome-wide meta-analysis in Japanese populations identifies novel variants at the TMC6–TMC8 and SIX3–SIX2 loci associated with HbA1c
Scientific Reports (2017)