Abstract
By regulating access to the genetic code, DNA supercoiling strongly affects DNA metabolism. Despite its importance, however, much about supercoiled DNA (positively supercoiled DNA, in particular) remains unknown. Here we use electron cryo-tomography together with biochemical analyses to investigate structures of individual purified DNA minicircle topoisomers with defined degrees of supercoiling. Our results reveal that each topoisomer, negative or positive, adopts a unique and surprisingly wide distribution of three-dimensional conformations. Moreover, we uncover striking differences in how the topoisomers handle torsional stress. As negative supercoiling increases, bases are increasingly exposed. Beyond a sharp supercoiling threshold, we also detect exposed bases in positively supercoiled DNA. Molecular dynamics simulations independently confirm the conformational heterogeneity and provide atomistic insight into the flexibility of supercoiled DNA. Our integrated approach reveals the three-dimensional structures of DNA that are essential for its function.
Similar content being viewed by others
Introduction
The structure of the B-form DNA double helix has been known for over 60 years1, yet DNA metabolism requires deviations, sometimes extreme, from this canonical form2,3,4. Studies of DNA are typically performed on short, linear DNA fragments that cannot be supercoiled; therefore our understanding of DNA is incomplete. In most organisms DNA is maintained in a negatively supercoiled (underwound) state. Positively supercoiled (overwound) DNA is transiently generated during DNA replication and transcription, and, if not promptly relaxed, inhibits these processes5. Electron microscopy6,7,8,9,10,11,12,13 (including electron cryo-microscopy6,8,11,12,13) and atomic force microscopy10,14 have previously provided significant insight into the structure of negatively supercoiled DNA. Here we build on those earlier studies by covering a broader range of supercoiling, including positive supercoiling.
Using a combination of approaches we here show that supercoiling causes DNA to adopt large conformational variability. Unpaired bases and localized distortions contribute to this variability both by relieving torsional strain and by providing flexible hinges. Positive and negative supercoiling are accommodated differently. The biological implications of these findings are discussed.
Results
336 bp minicircles accommodate a wide range of supercoiling
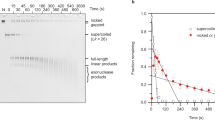
Minicircles containing 336 bp were selected for this study because they are representative of the supercoiled DNA loops found in nature15,16,17,18. Furthermore, these minicircles are small enough to allow isolation of suitable amounts of individual topoisomers (Fig. 1a), yet large enough to yield an ample spread of ten unique topoisomers (Supplementary Fig. 1). In its relaxed state, the two strands of a 336 bp DNA circle wrap around each other 32 times. This number is known as the linking number (Lk). The other topoisomers deviate (ΔLk) from the Lk of the relaxed topoisomer (Lk=32, ΔLk=0) in steps of 1. We generated and isolated six different negatively supercoiled minicircle topoisomers (Lk=31 through 26, ΔLk=−1 through −6), three different positively supercoiled topoisomers (Lk=33 through 35, ΔLk=+1 through +3), relaxed (Lk=32, ΔLk=0), nicked and linearized minicircles (Fig. 1a, Supplementary Table 1).
(a) Individual 336 bp minicircle topoisomers were isolated and analysed by polyacrylamide gel electrophoresis in the presence of 10 mM CaCl2. Mr: 100 bp DNA ladder, L: minicircle linearized by EcoRV, N: minicircle nicked by Nb.BbvCI. (b) Projections of cryo-ET subtomograms of hydrated 336 bp DNA minicircles of the Lk=34 topoisomer. (c) Commonly observed shapes were open circle, open figure-8, figure-8, racquet, handcuffs, needle, and rod, each of which are shown in orthogonal views. (d) Other shapes observed, especially in the more highly supercoiled topoisomers. (e) Shape frequency distribution plot for each topoisomer population (n=number of minicircles analysed). A weighted average for each topoisomer, approximating the average degree of compactness, is denoted by the black triangle. The weighted average was calculated by assigning each conformation a value that increased in line with compactness. Open circles were given a value of 1, open figure-8 s a value of 2, figure-8 s as a value of 3, and so on. The relative fraction of each was subsequently used to determine the average degree of compactness. Lk, ΔLk and superhelical density (σ) for each topoisomer are shown (see Supplementary Note 1).
All nine of the supercoiled minicircles were subjected to and relaxed by human topoisomerase IIα (htopoIIα) (Supplementary Fig. 2). This relaxation demonstrated the biological activity of the minicircles, confirmed their Lk designations, and verified that there was no cross-contamination among topoisomers. Because htopoIIα relaxes in characteristic steps of two Lk, all of the minicircles with even Lk relaxed to ΔLk=0. The odd-numbered Lk topoisomers relaxed to a mixture of ΔLk=−1 and ΔLk=+1.
Wide variety of minicircle DNA conformations
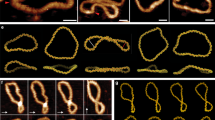
Electron cryo-tomography (cryo-ET) was used to obtain three-dimensional (3D) structures of the different minicircles embedded in vitreous ice. Before freezing, the purified topoisomers were either incubated on ice or at room temperature for at least 15 min in an attempt to allow the DNA to reach conformational equilibrium. Specimen vitrification19 occurs at a rate of freezing (106 °C per second) that should be fast enough to preclude temperature-dependent structural alterations20. Projection images were obtained with an electron microscope by incrementally tilting the specimen stage. These images were subsequently reconstructed into 3D volumes (tomograms) containing the minicircles (Supplementary Movie 1). Thus, these data represent snapshots of the DNA molecules at the instant of freezing. We computationally generated traces of 336-bp minicircle DNA backbones and fitted these to the observed densities in the tomograms, confirming that each subvolume represented a single 336 bp minicircle (Fig. 2a,b, Methods). In addition, as a control, we visualized double-length minicircles (672 bp) and computationally fitted a 672 bp backbone to the observed densities, confirming their length and thus providing additional validation of the approach (Fig. 2c).
(a) Docking of 336 bp traces into the cryo-ET densities of open circles. Traces (purple lines) were generated by docking circular strings of length 336 bp into the density maps. Each trace was then used to isolate the minicircles (grey surfaces) from the cryo-ET density maps. (b) Docking of 336 bp traces into the cryo-ET tomograms of writhed minicircles following the same protocol as for the open circles. (c) Docking of double-length (672 bp) traces into the cryo-ET tomograms following the same protocol as for 336 bp.
A broad mixture of 3D conformations was observed for each purified 336-bp minicircle topoisomer (Fig. 1b,e). Given the rigidity of DNA at this short length21, it is remarkable that the DNA was able to contort into such a wide variety of conformations. The heterogeneity observed indicates that the DNA structure is in dynamic equilibrium, driven by both the torsional stress of supercoiling and by Brownian motion13. These results illustrate the power of cryo-ET to visualize individual molecules in solution and capture their conformational variability. Furthermore, cryo-ET data contained sufficient detail to visualize that, as expected, only right-handed crossovers were observed in negatively supercoiled DNA and only left-handed crossovers were observed in positively supercoiled DNA.
Most of the observed minicircle conformations could be classified into the following empirical categories, in order of increasing compaction: open circle, open figure-8, figure-8, racquet, handcuffs, needle, and rod (Fig. 1b,c). Examples of the handcuffs and needle conformations are shown in Supplementary Movie 2 and 3. It is interesting to note that minicircles in topologically distinct topoisomers sometimes adopted the same general conformation (for example, rods found in the ΔLk=−1 and ΔLk=−6 topoisomers appear similar). In these cases, DNA supercoiling must be accommodated in different ways that result in the same general 3D shape. Some minicircles, especially those with larger ΔLk (either negative or positive) adopted alternative shapes (‘other’ in Fig. 1d).
Deviations from relaxed DNA may be manifested as changes in twist, the coiling of the DNA about the helical axis, or writhe, the coiling of the double helices about each other. Changes in twist result in torsional strain whereas changes in writhe result in bending strain. Because we observed multiple DNA conformations in supercoiled DNA—ranging from open to highly writhed—different degrees of twist and writhe must simultaneously exist within the same topoisomer population. DNA bending, and thus writhe, is thought to be more difficult to accommodate for short DNA lengths. Minicircles twice the length (672 bp) with equivalent supercoiling (ΔLk=−4) to the ΔLk=−2, 336 bp minicircle all appeared highly writhed (Fig. 2c). In comparison the ΔLk=−2, 336 bp topoisomer displayed both open and writhed conformations (Fig. 1e). Conversely, smaller minicircles of 94–158 bp have no appreciable writhe11,12,13. Minicircles (336 bp) are, therefore, an ideal size for exploring the relationship between twist and writhe.
As mentioned above, linear DNA is rigid in the aforementioned lengths. Theory therefore predicts that very small circles should be perfectly round. Significant deviations from a perfect circle will require non-uniform distribution of bending along the DNA length. Very small circles already have considerable bending strain. Localized variations in bending should therefore be energetically unfavourable. Ellipticity was measured previously in 94–158 bp DNA minicircles and averaged between 1.1 and 1.5 (refs 11, 12, 13). Increased ellipticity was attributed to the appearance of hyperflexible kinks within the DNA13. Thus ellipticity is one measure of conformational variability. To compare our data to those previous studies, we measured ellipticity in a subset of our observed open circles. Significant numbers of minicircles with an open circle shape were found in the following topoisomer populations: ΔLk=−2, 0, +1, +2, +3, and nicked. The measured ellipticity values ranged from 1.1 to 2.6. This larger deviation from circularity is attributable to the longer length of our 336 bp minicircles. We also observed potential differences with supercoiling in the dimensions of the open circles (Supplementary Fig. 3).
The conformations adopted by each topoisomer are shown as frequency distribution plots (Fig. 1e). A weighted average of the shape frequency distribution (denoted by arrowheads on each plot) approximates the average degree of compactness. Similarly, electrophoretic mobility provides a measure of relative compaction22. By both measures increased negative or positive supercoiling leads to a shift in the distribution towards more compact shapes. To provide a more quantitative measure of compactness, radius of gyration values were measured from the cryo-ET density maps (Fig. 3a, middle). These values were consistent with the electrophoretic mobilities (Fig. 3a, left).
(a) Left, distance each topoisomer migrated during polyacrylamide gel electrophoresis, measured from the well to the centre of the band (Fig. 1 a), relative to the migration of the linearized 336 bp minicircle. Data shown are the mean values from three separate gels run under identical conditions. Middle, average radius of gyration values obtained from cryo-ET density maps for each topoisomer (n=23, 78, 40, 60, 47, 56 and 159 for topoisomers ΔLk=−3, −2, −1, 0, 1, 2 and 3, respectively). Right, radius of gyration (averaged over time) in continuum solvent MD simulations for each topoisomer. Error bars for each of the three graphs represent s.d. values. (b) Comparison of cryo-ET data and the equivalent conformations as observed in MD simulations. Examples from negatively supercoiled (ΔLk=−2) and positively supercoiled (ΔLk=+1) topoisomers are shown. MD simulation data are depicted as double-stranded DNA backbone traces.
Molecular dynamics simulations provide atomistic insight
To further understand the conformational fluctuations of the topoisomers, we performed molecular dynamics (MD) simulations (Supplementary Movie 4 and 5; Supplementary Tables 2 and 3). Generalized Born continuum solvent simulations, which, in the absence of viscous damping enable rapid configurational sampling in a relatively short computational time, revealed a wide variety of conformations for each topoisomer. The simulation results suggest that each supercoiled topoisomer undergoes large fluctuations in writhe (Supplementary Movie 4 and Supplementary Fig. 4) and, hence, in the level of compaction. The radius of gyration values, averaged over the MD simulations, showed the same trends with changing superhelical density as the gel electrophoretic mobility and the radius of gyration values extracted from the cryo-ET data (Fig. 3a). Conformations that were observed in the cryo-ET data were also observed in the MD simulations (Fig. 3b), providing insight into how conformations may interchange. The consistency between structural and computational results established confidence for the observation of simultaneous co-existence of multiple conformations of each topoisomer.
Across all levels of supercoiling, differences were observed for negative versus positive ΔLk. Although the ΔLk=−1 and ΔLk=+1 topoisomers might be expected to migrate with similar electrophoretic mobility, ΔLk=−1 migrated much faster on the gel (Fig. 1a). In addition, cryo-ET data revealed that the ΔLk=−1 topoisomer adopted a spread of conformations that included predominantly compact forms (Fig. 1e). The ΔLk=+1 adopted mostly open conformations, similar to the nicked and relaxed minicircles (Fig. 1e). The presence of a small fraction (∼10%) of compact forms may explain why, on average, the ΔLk=+1 topoisomer migrated slightly farther on the gel than the nicked or relaxed minicircles.
Extending the comparison to ΔLk=−2 and ΔLk=+2, we observed broad distributions of conformations, such that every shape category described in Fig. 1c was seen. Overall, the distribution trends for the ΔLk=−2 and ΔLk=+2 were similar. One noteworthy difference, however, was in the relative proportions of the figure-8 and racquet conformations. Racquets were observed ∼3-fold more frequently than figure-8 s for the ΔLk=−2 topoisomer; figure-8 s were ∼5-fold more frequent than racquets for ΔLk=+2. This observation implied structural differences between the two topoisomers, which we verified biochemically as described below.
Further underwinding (ΔLk=−3, −4 and −6) resulted in an additional shift in the distribution toward more compact shapes observed in the cryo-ET data and a concomitant increase in electrophoretic mobility. The increase in electrophoretic mobility between consecutive topoisomers was more pronounced for positive than for negative supercoiling (Fig. 1a). A similar trend was observed for the shift in the conformational distribution between consecutive topoisomers (Fig. 1e). For ΔLk=+3, 39% of the minicircles adopted unusual shapes (‘other,’ Fig. 1d), which were not observed in the ΔLk=+2 population (Fig. 1e), indicating that a sharp structural transition occurs between these two topoisomers.
Probing for base-pair disruptions and localized denaturation
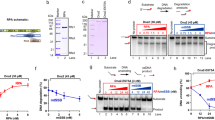
Many of the observed 3D conformations appear to contain sharply bent or kinked DNA. One way that sharp bending may be facilitated is through localized distortions and disruptions of the helix. To probe for and quantify such helix disruptions, we used nuclease Bal-31. Bal-31 has endonuclease function on exposed, unpaired DNA bases (for example, kinks, nicks, gaps, single-stranded regions and B–Z junctions)23,24. All underwound topoisomers and the most overwound ΔLk=+3 topoisomer had some exposed, unpaired bases, as measured by their Bal-31 sensitivity (Fig. 4a). The negatively supercoiled ΔLk=−2, −3, −4 and −6 topoisomers were linearized within the first minute of Bal-31 incubation and subsequently degraded (Fig. 4b). The ΔLk=−1 topoisomer was cleaved at a much slower rate, requiring 20 min to degrade (Fig. 4b). Consistent with their Bal-31 sensitivity, the electrophoretic mobility of the negatively supercoiled topoisomers shifted when incubated with glyoxal, a small molecule that traps exposed bases (Supplementary Fig. 5). The relaxed (ΔLk=0) and ΔLk=+1 topoisomers resisted Bal-31 digestion, and the ΔLk=+2 topoisomer was a very poor substrate (Fig. 4b). Considering that positive supercoiling should inhibit strand separation, it was surprising that the ΔLk=+3 topoisomer was efficiently cleaved by Bal-31 (Fig. 4a) and this topoisomer was almost completely degraded within an hour (Fig. 4b). Single molecule manipulation studies previously uncovered a structural variation of DNA when it was extremely overwound25. The researchers proposed that overwinding may result in an inside-out DNA conformation with the backbones wrapped around each other on the inside and unpaired bases on the outside of the helix. Chemical probing confirmed the presence of unpaired bases. This DNA conformation, named Pauling DNA (P-DNA), was only detected when the DNA was under high tension and writhe was suppressed25. The Bal-31 sensitivity of ΔLk=+3 coincides with the dramatic structural changes observed by cryo-ET for this topoisomer (Fig. 1e). Although the presence of exposed bases may imply P-DNA, there are alternative explanations for Bal-31 sensitivity. A probable explanation for the exposed bases is denaturation resulting from sharp bending. Indeed, sharp bending is a feature of the highly writhed conformations of this topoisomer (see below).
(a) Minicircle DNA incubated with nuclease Bal-31. Over time, samples were removed, quenched by the addition of stop buffer and the products analysed by polyacrylamide gel electrophoresis. Mr: 100 bp DNA ladder, L: linearized 336 bp DNA, N: nicked 336 bp minicircle. (b) Graphic representation of the data shown in (a) Fitted lines are for visualization purposes only. (c) MD simulation of the ΔLk=−3 topoisomer in explicit solvent. Splayed bases were found at a sharp bend of a needle conformation. This may be a potential atomistic explanation for Bal-31 susceptibility of negatively supercoiled topoisomers.
To provide an atomistic interpretation of the cryo-ET density for each topoisomer and to explore how localized distortions may alter the structural properties of DNA, we performed explicitly solvated MD simulations (Supplementary Movie 6–9). Because the inclusion of solvent molecules in the calculations slows down configurational sampling, these simulations were performed starting from conformers most commonly observed in the continuum solvent MD. The explicit solvent simulations allowed increased conformational diversity and could be directly fitted to the cryo-ET density maps (Fig. 3b and Supplementary Movie 10–13). Whereas restraints were imposed on the hydrogen bond interactions between complementary base pairs in the simulations in implicit solvent, these restrictions were removed for simulations in explicit solvent. This removal of restraints allowed single-stranded regions to form within the minicircles under sufficiently high levels of torsional or bending stress. Accordingly, in simulations of the ΔLk=−3 topoisomer, a kink emerged at the apex of a highly writhed minicircle, where the hydrogen bonding between complementary base pairs was disrupted (Fig. 4c). Similar base-pair disruptions observed by MD simulations have been reported for bent DNA26,27 and for underwound linear DNA28. These computational results are consistent with the experimental observation that the ΔLk=−3 topoisomer is under a high degree of torsional strain, but additionally predicts that base-pair separation may occur at bent apices. Vologodskii and co-workers found that very small DNA minicircles (64–65 bp) were susceptible to Bal-31 cleavage, which was attributed to kinking resulting from the inherent bending strain29. Base-pair disruption has been observed in MD simulations of positively supercoiled DNA, either because of the formation of P-DNA28 or because of sharp bending in very small DNA circles27.
The propensity for base-pair separation depends on DNA sequence30,31. We used the WebSIDD algorithm32 to predict base-pair separation in our 336 bp minicircle, which identified a short region within attR to have a higher probability of duplex destabilization. Because it was very susceptible to Bal-31, the ΔLk=−6 topoisomer was probed. In contrast to the prediction, Bal-31 predominantly cleaved ∼180° away from the anticipated destabilization sequence (Fig. 5). Increased flexibility brought about by localized denaturation at the Bal-31-susceptible site should enable the DNA to be sharply bent. The majority of conformations observed for the ΔLk=−6 topoisomer were sharply bent or kinked (for example, racquets and rods). Kinking at this first site may induce kinking at the site diametrically opposite. Similar cooperative effects between distant sites were seen by Stasiak and co-workers, who observed sequential, cooperative kinking in torsionally strained minicircles at sites located ∼180° apart along the DNA circumference13. An additional influence of the location of Bal-31-susceptible sites may be sequence-dependent DNA flexibility and curvature. Sequences with increased flexibility and/or curvature are preferentially localized to superhelical apices33 where the DNA is most sharply bent. A short region in attR is known to have modest intrinsic curvature34,35. This curvature may be preferentially located at a superhelical apex36, thereby positioning the sequence located ∼180° apart along the DNA circumference at the diametrically opposite superhelical apex. Localized denaturation may be necessary to accommodate the sharp bending at the apex, thereby generating a Bal-31-susceptible site. The kink defect observed by MD at a bent apex (Fig. 4c) is relatively close to the Bal-31 cleavage site (Fig. 5b), consistent with the hypothesis that Bal-31-susceptible sites are localized to the superhelical apices.
To determine whether Bal-31 cleavage occurs at multiple sites or at a preferred site, the ΔLk=−6 topoisomer was cleaved with Bal-31 and various restriction enzymes. (a) Products were separated by agarose gel electrophoresis. Left, (lanes 1–5), control reactions, mc336 (approximately equal mixture of ΔLk=−2 and ΔLk=−3 topoisomers) with combinations of the various restriction enzymes (as indicated) to generate fragments of known DNA lengths. Right, (lanes 6–9), ΔLk=−6 topoisomer cleaved first with Bal-31, followed by a restriction enzyme (as indicated). Mr1: 100 bp DNA ladder, Mr2: Low molecular weight DNA ladder. (b) Map of the minicircle sequence showing the positions of the restriction enzymes used, the estimated location of Bal-31 cleavage (with parentheses indicating the range), and the location of the observed base-pair breaking in MD simulation of the ΔLk=−3 topoisomer.
Discussion
Our data collectively demonstrate that supercoiled DNA is able to adopt a wide variety of conformations, the proportions of which depend upon the level of supercoiling. Each topoisomer appears to migrate as a single discrete electrophoretic band (Fig. 1a), suggesting that each individual DNA molecule fluctuates among the different conformations seen by cryo-ET. If there were no interchange among conformations, we would expect multiple discrete bands (representing the different meta-stable conformations), a smear, or a more diffuse band. MD simulations provided further evidence of the dynamic nature of supercoiled DNA and insight into how the conformations may interconvert.
Each topoisomer displayed a wide variety of conformations but the relative proportion of these depended on Lk. Because of rapid vitrification, the structures observed should represent a snapshot of the highly dynamic DNA molecule. More highly writhed conformations were observed with increasing negative or positive supercoiling. These trends show how supercoiling drives the dynamic equilibrium and determines how frequently and for how long each minicircle adopts a particular conformation. Somewhat surprisingly, a small number of open circles were observed even in the ΔLk=−6 topoisomer. For these open circles the ΔLk must be partitioned almost entirely into twist.
Sensitivity to Bal-31 cleavage revealed the presence of exposed bases when the DNA was negatively supercoiled, and also when highly positively supercoiled. On one hand, localized Bal-31-susceptible distortions create flexible hinges that allow DNA to assume conformations that would be energetically unfavourable if the bases remained paired. On the other hand, extreme writhe and concomitant bending at the apices lead to kinking and localized distortions, even in positively supercoiled DNA. More extensive denaturation relieves torsional strain, one potential explanation for why a small number of open circles are observed in the ΔLk=−6 topoisomer. The release of torsional strain through denaturation may also explain why more open circles are observed in the ΔLk=−2 topoisomer than the ΔLk=−1 topoisomer (Fig. 1e). The interplay between localized denaturation and the sharp bending required to achieve the observed conformations is summarized in Fig. 6. Mapping of the cleavage site of Bal-31 suggested that sharp bending at one location may in turn lead to localized distortions at a site located ∼180° away along the perimeter of the minicircle, similar to the cooperative kinking model13.
Comparison of smooth and sharp bending and the effect of localized denaturation. Images represent a more detailed view of the local structure at the bend. For smooth bending, writhe-mediated bending is regular with bending strain more evenly distributed. Base flipping may generate flexible hinges, allowing DNA to bend more sharply or kink. Alternatively, writhe-mediated sharp bending may lead to disruption of base pairs, even in positively supercoiled DNA. More extensive denaturation may release torsional strain and allow DNA to adopt more open conformations. Denaturation bubbles also provide a flexible joint allowing DNA to kink.
Our results provide a glimpse into the dynamics and structure of DNA in vivo not captured by experiments using short, linear DNA. The situation described in our paper is most comparable to unconstrained DNA, which constitutes a significant fraction of the genomes of Escherichia coli, humans, and presumably other organisms37,38,39. Supercoiling, even in the constrained portions of chromosomes, modulates bound architectural proteins. In turn, the bound proteins influence the conformational variability of supercoiling. A protein may be able to exploit the conformational variability of supercoiled DNA by transiently stabilizing one of the conformations, allowing switching between active and inactive states. In actively replicating and transcribing cells, supercoiling fluctuation serves multiple purposes. Positive supercoiling ahead of RNA polymerase facilitates histone dissociation during chromatin remodelling40 and important regulatory elements, such as the FUSE element in the c-myc promoter3,41 or the putative hairpin in the N4 promoter of vRNAP42 are tunable by supercoiling.
Six decades after the elucidation of its double helical structure, DNA continues to surprise us by revealing new information. Our cryo-ET, biochemical, and computational studies show the astounding versatility and dynamism of DNA depending on the degree of supercoiling. DNA simultaneously exists in a largely inactive B-form with bases tucked in and protected and an active, highly varied structure with exposed bases. Our data provide relative comparisons of supercoiling-dependent twisted, writhed, curved, and kinked conformations and associated base exposure. Each of these structural features may be differentially recognized by the proteins, nucleic acids, and small molecules that modulate DNA metabolic processes.
Methods
Chemicals and reagents
ATP, dithiothreitol (DTT), DNase I, ethidium bromide, glyoxal and RNase A were purchased from Sigma Aldrich (St Louis, MO). Acrylamide, ampicillin, chloroform, isopropyl beta-D-thiogalactoside, sodium chloride and sodium citrate were purchased from Fisher Scientific (Pittsburgh, PA). BbvCI, EcoRV, Nb.BbvCI, NdeI, Nuclease Bal-31, T4 DNA Ligase, low molecular weight DNA ladder and 100 bp DNA ladder were purchased from New England Biolabs (Ipswich, MA). Proteinase K was purchased from Roche Molecular Biochemicals (Mannheim, Germany). All other chemicals were purchased from VWR International (West Chester, PA).
Expression and purification of HMfB
The expression construct pKS323 (ref. 43) was transformed into CGSC-10851 and cells were grown at 37 °C to an OD600=0.4 in Luria–Bertani broth supplemented with 100 μg ml−1 ampicillin. HMfB expression was induced by the addition of 0.4 mM isopropyl beta-D-thiogalactoside and expression continued for 16 h. Cells were harvested by centrifugation, resuspended (2 ml per gram of cells) in 50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 2 mM Na2HPO4 and lysed by two passages through a French pressure cell press at 20,000 psi. Cell debris was removed by centrifugation at 30,000g for 30 min. Cleared lysates were transferred to new tubes and centrifuged for another hour at 30,000g. The supernatant, after addition of 5 mM MgCl2 and 0.1 mM phenylmethylsulfonyl fluoride, was treated with 20 μg ml−1 DNase I at 37 °C for ∼3 h. NaCl was added to a final concentration of 3 M and the mixture was heated slowly to 80 °C for 10 min, resulting in precipitation of native E. coli proteins. The mixture was cooled to room temperature, filtered through several layers of cheesecloth, filtered though a 0.45 μM filter, dialyzed overnight at 4 °C against HMfB storage buffer (50 mM Tris-HCl, pH 8.0, 333 mM sodium citrate) and stored at 4 °C. This ‘crude extract’ was used in the ligations without further purification.
Generation and purification of minicircle topoisomers
Plasmid pMC336, constructed by deleting three base pairs from pMC339-BbvCI (ref. 14) using the QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA), generates both the 336 bp and 672 bp minicircles. Plasmid sequences were confirmed by DNA sequencing (Lonestar Labs, Houston, TX). DNA minicircles (minivectors™) were purchased from Twister Biotech (Houston, TX). Minicircle DNA was nicked at a single site using nicking endonuclease Nb.BbvCI according to the manufacturer’s protocol. The reaction was subsequently incubated at 80 °C to inactivate the nicking endonuclease, extracted with 25:24:1 phenol:chloroform:isoamyl alcohol, extracted with chloroform, precipitated with ethanol, and resuspended in TE buffer (10 mM Tris-HCl, pH 8.0, and 1 mM disodium Ethylenediaminetetraacetic acid (EDTA)). Negatively supercoiled topoisomers were generated using ethidium bromide as described14. Positively supercoiled topoisomers were prepared using HMfB crude extract. The method of generating supercoiling did not influence the structure of the minicircles (Supplementary Fig. 1b). Optimal HMfB:DNA ratios were empirically determined for each HMfB preparation. Nicked minicircle DNA was incubated in the presence of HMfB crude extract for 15 min in 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM ATP and 10 mM DTT. T4 DNA ligase was added and the reaction mix was incubated for 4 h at 32 °C. Disodium EDTA (20 mM) was added to quench ligation, followed by the addition of RNase A (50 μg ml−1 ) and the mixture was incubated at 37 °C for 30 min. Proteinase K (50 μg ml−1 ) was added and the reaction was incubated at 45 °C for 1 h in the presence of 1% SDS. The mixture was concentrated and desalted using an Amicon centrifugal filter (EMD Millipore, Billerica, MA), precipitated with ethanol, and the DNA was resuspended in TE buffer (pH 8.0).
Individual minicircle topoisomers were separated by electrophoresis in 5% polyacrylamide gels (acrylamide:bis-acrylamide=29:1) at 125 V (∼6 V cm−1 ) for 8 h in the presence of Tris-acetate buffer (pH 8.2) containing 10 mM CaCl2 (for negatively supercoiled topoisomers), 1 mM disodium EDTA (for positively supercoiled topoisomers) or 1 mM disodium EDTA with 2 μg ml−1 ethidium bromide (for the relaxed, Lk=32, topoisomer). Preparative gels were stained with ethidium bromide. DNA was electroeluted from the gel slices at 80 V for ∼16 h at room temperature in D-tube dialyzers (Novagen, Madison, WI) in 40 mM Tris-acetate buffer (pH 8.2) with 1 mM disodium EDTA. Electroeluted DNA was extracted thrice with 1-butanol to remove residual ethidium bromide, extracted with chloroform, precipitated with ethanol, resuspended in 10 mM Tris-HCl, pH 8.0, and 0.1 mM disodium EDTA, desalted using an Amicon 0.5 ml centrifugal filter, precipitated again in ethanol, and resuspended in 10 mM Tris-HCl, pH 8.0, and 0.1 mM disodium EDTA. DNA concentrations were determined using a Nanodrop spectrophotometer (Thermo Scientific, Wilmington, DE). DNA samples were analysed by electrophoresis through 5% polyacrylamide gels (acrylamide:bis-acrylamide=29:1) in Tris-acetate buffer (pH 8.2) containing 10 mM CaCl2 at 125 V (∼6 V cm−1 ) for ∼8 h. Buffer was recirculated continuously throughout electrophoresis. Gels were stained with SYBR Gold (Life Technologies, Grand Island, NY) and visualized using a FOTO/ANALYST Investigator imaging system (Fotodyne, Hartland, WI) with quantification using the TotalLab software (TotalLab, Newcastle, UK).
Relaxation assay with hTopoIIα
Minicircle topoisomers (336 bp, 25 ng) were incubated with hTopoIIα (10:1 DNA:enzyme molar ratio) in 10 mM Tris-HCl, pH 7.9, 175 mM KCl, 0.1 mM disodium EDTA, 5 mM MgCl2 and 2.5% glycerol in a total reaction volume=20 μl. ATP was added to a final concentration of 1 mM to start relaxation, and the reaction mixes were incubated at 37 °C for 20 min. Reactions were stopped by the addition of 3 μl of 50 mM disodium EDTA and 5% SDS (final concentration, 6.5 mM and 0.7%, respectively). Proteinase K was added (final concentration=1 μg μl−1 ) and the mixtures were incubated for 30 min at 45 °C. DNA topoisomers were resolved on a 5% polyacrylamide gel (acrylamide:bis-acrylamide=29:1) in 40 mM Tris-acetate containing 10 mM CaCl2 for 60 V for ∼16.5 h. Buffer was recirculated continuously throughout electrophoresis. Gels were destained in 40 mM Tris-acetate containing 10 mM disodium EDTA, followed by subsequent staining with SYBR Gold and visualized using a FOTODYNE FOTO/analyst investigator imaging system.
Electron cryo-tomography
DNA samples (100 ng μl−1) in Tris-acetate buffer (pH 8.2) supplemented with 10 mM CaCl2 were applied onto glow-discharged 200-mesh copper Quantifoil R 1.2/1.3 holey carbon grids (Quantifoil Micro Tools GmBH, Großlöbichau, Germany) that had been pretreated with 15 nm gold-BSA fiducials. The grids were blotted and vitrified using a Mark IV Vitrobot (FEI, Hillsboro, OR), and stored in liquid nitrogen. Tilt series were collected on a JEM2200FS 200 kV electron microscope (JEOL, Tokyo, Japan) equipped with a field emission gun, energy filter (set to 20 eV) and a 4 k by 4 k CCD camera (Gatan, Pleasanton, CA). The specimen was maintained at −175 °C in a Gatan Model 626 cryo-holder. The microscope settings were spot size=5, condenser aperture=100 μm and objective aperture=60 μm. Images were collected at ∼25,000x magnification, typically from −60° to +60° nominal tilt, in 3° (or in one case 10°) increments, with intended defocus of 5−7 μm using SerialEM44. Final sampling was 4.52 Å pixel−1. Tomograms were reconstructed in IMOD45 and visualized in 3dmod. Individual DNA minicircle topoisomers were identified by eye in 3dmod and computationally extracted. A Gaussian low-pass filter was applied to each subvolume before visualization in UCSF Chimera46.
Within each population, minicircles were classified visually into categories by at least two individuals. Differences among categories were sometimes subtle. The difference between the racquet and the needle, for example, was only in the relative sizes of the loop and the handle. Variations within the needle populations were in the size of the loop and amount of bending of the minicircle. Minicircles were classified as ‘Other’ if they did not conform to the shapes shown in Fig. 1c. Five of the most prominent alternative configurations are shown in Fig. 1d; however, we also observed densities that appeared to be minicircles, but their shapes were difficult to decipher.
Computational tracing of DNA minicircle
First, cryo-ET density maps were normalized, low-pass filtered, and disconnected noise was removed computationally47. Initially, five connected vertices in a closed loop were placed around each minicircle. A score was calculated including the density value at the vertices, the length of each edge, and the angle between the edges. Each vertex was then iteratively moved in the direction of the density gradient to optimize the score, with the length and angle terms acting as restoring forces to cause the shape to remain as much like an open loop as permitted by the density map. After this initial optimization, the number of vertices was doubled by adding a point in the middle of each edge. The set of vertices was subsequently optimized in the same way and the procedure was repeated once more with 20 vertices. The convergence of the algorithm was verified using multiple random starting locations and assessing the similarity of results. Relative axis lengths were computed from the coordinates of the final polygon for each minicircle.
To compute the radius of gyration, the raw density for each minicircle was extracted by directly including any voxels within 45 Å of the fit path, then damped to a Gaussian decay extending to a distance of 180 Å. The radius of gyration was then calculated from the extracted subvolume using the position and density of each voxel, making no direct use of the fit vertices.
Molecular dynamics simulations
Starting structures of 336-bp minicircle DNA with different helical twists (corresponding to seven Lk values of 27–33) were built using the NAB module implemented in AMBERTOOLS11 (ref. 48). The AMBER99 force field with parmbsc0 corrections for the α and γ dihedral parameters49 and a correction for χ (glycosidic bond) parameters50 was used to describe the DNA. Following a multistage equilibration protocol previously described51, these starting structures were subjected to 20 ns of implicitly solvated MD using the Tsui and Case Generalized Born/Solvent Accessible area (GB/SA) method52 at 300 K and 100 mM monovalent salt concentration, which is roughly equivalent to 10 mM CaCl2 (ref. 53), with the long-range electrostatic cutoff set to 100 Å. Restraints were imposed on the hydrogen bonding interactions between complementary base pairs. To determine the superhelical density of each of the seven topoisomers, we identified the relaxed topoisomer (which must be the least compact minicircle) by calculating the radius of gyration (Fig. 3a). In the simulations, ΔLk=0 actually corresponds to an Lk of 30 (reflecting the known underestimation of relaxed DNA twist in the AMBER forcefield51).
We performed these initial MD simulations with a continuum representation of the solvent to more rapidly explore conformational space in the absence of any frictional drag from collisions with water molecules. Discarding the first 5 ns for equilibration, the calculated average writhe for each topoisomer did not change by more than 8% for each topoisomer when the simulations were extended from 15 to 20 ns; consequently we considered the writhed to be adequately sampled by the implicitly solvated simulations (Supplementary Fig. 4). We then performed clustering analysis using the average linkage algorithm54 in the PTRAJ package in AMBERTOOLS11. A representative structure of the most populated cluster was chosen and solvated in 100 mM Na+ and Cl- counterions55 and a TIP3P water box56, and 10 ns MD simulation runs were performed using the GROMACS 4.5 program57 with standard MD protocols (Supplementary Information). By comparing the maximum relative speeds of the centres of mass of two DNA bases located on opposite sides of the minicircle in implicit and explicitly solvated simulations, we estimated that inclusion of the frictional term from solvent/solute interactions retards the dynamics of the DNA by about an order of magnitude. This difference suggests that the timescale required for interconversion among minicircle conformers lies between 100 ns and a microsecond. We then calculated RMSDs between the simulated atomistic models in explicit solvent and the ensemble of computational traces obtained from cryo-ET, and selected the four MD models with the lowest RMSD for direct comparison with the cryo-ET density maps (Fig. 3 and Supplementary Movie 10–13).
Nuclease Bal-31 assay
Minicircle DNA (300 ng in 60 μl final volume) was incubated with 0.3 units of nuclease Bal-31 at 30 °C in 20 mM Tris-HCl, pH 8.0, 600 mM NaCl, 12 mM MgCl2, 12 mM CaCl2 and 1 mM disodium EDTA. At 1, 10, 20 and 60-minute intervals, 10 μl (50 ng) samples were removed, quenched by addition of an equal volume of stop buffer (50 mM Tris-HCl, pH 8.0, 100 mM disodium EDTA, 10% glycerol, 200 μg ml−1 proteinase K), followed by incubation at 45 °C for 30 min to degrade Bal-31. Products were analysed by electrophoresis through 5% polyacrylamide gels (acrylamide:bis-acrylamide=29:1) in Tris-acetate buffer (pH 8.2) containing 1 mM disodium EDTA. Gels were run for 4 h at 125 V (∼6 V cm−1). Buffer was recirculated continuously throughout electrophoresis. Gels were stained with SYBR Gold and visualized using a FOTO/ANALYST Investigator imaging system with quantification using the TotalLab software.
Mapping Bal-31 cleavage
Minicircle DNA (7.5 μg of the ΔLk=−6 topoisomer in 3 ml final volume) was incubated with 7.5 units of nuclease Bal-31 at 30 °C in 20 mM Tris-HCl, pH 8.0, 600 mM NaCl, 12 mM MgCl2, 12 mM CaCl2 and 1 mM disodium EDTA. After 1 min the reaction was quenched by the addition of disodium EDTA (50 mM final concentration) and incubated with proteinase K (100 μg ml−1 ) at 45 °C for 1 h. The reaction was concentrated and desalted using an Amicon centrifugal filter. Full length linearized DNA was isolated on 5% polyacrylamide gels (acrylamide:bis-acrylamide=29:1) at 125 V (∼6 V ) for 4 h in the presence of Tris-acetate buffer (pH 8.2) containing 1 mM disodium EDTA. The preparative gel was stained with ethidium bromide and the DNA recovered from the gel as described above.
Bal-31 linearized DNA (100 ng) was incubated with EcoRV, BbvCI or NdeI according to the manufacturer’s protocol. Control reactions were performed with minicircle DNA supplied by Twister Biotech, Inc. (an approximately equal mixture of ΔLk=−2 and ΔLk=−3 topoisomers). Products were analysed by electrophoresis on a 3% agarose gel (NuSieve 3.1 agarose, Lonza, Basel, Switzerland) at 100 V cm−1 for 2.5 h in Tris-acetate buffer (pH 8.2) containing 1 mM disodium EDTA. The gel was stained with SYBR Gold and visualized using a FOTO/ANALYST Investigator imaging system. Fragment sizes were determined by measuring distances migrated (measured from the well to the centre of each band using the TotalLab) and compared to a standard curve generated from the bands in the low molecular weight DNA ladder.
Glyoxal assay
Glyoxal was first deionized with AG-501-X8 mixed bed ion-exchange resin (Bio-Rad, Hercules, CA). 50 ng of minicircle DNA was incubated with 1 M glyoxal in 10 mM sodium phosphate, pH 7.0, for 16 h at room temperature. Control reactions were incubated in 10 mM sodium phosphate only. Samples were analysed by electrophoresis through 5% acrylamide gels (acrylamide:bis-acrylamide=29:1) in 10 mM sodium phosphate buffer (pH 7.0). Gels were run at 75 V for 6 h. Buffer was recirculated continuously throughout electrophoresis. Gels were stained with SYBR Gold and visualized using a FOTO/ANALYST Investigator imaging system. The diffuse spread of the negatively supercoiled topoisomers incubated with glyoxal precluded accurate quantitation of the data.
Additional information
Accession codes: Cryo-ET maps of representative minicircles of various shapes have been deposited into the EMDB under accession code EMD-6462.
How to cite this article: Irobalieva, R. N. et al. Structural diversity of supercoiled DNA. Nat. Commun. 6:8440 doi: 10.1038/ncomms9440 (2015).
Accession codes
Change history
29 October 2015
The original version of this Article contained an error in the spelling of the author Daniel J. Catanese Jr, which was incorrectly given as Daniel J. Catanese. This has now been corrected in both the PDF and HTML versions of the Article.
References
Watson, J. D. & Crick, F. H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 171, 737–738 (1953).
Fogg, J. M. et al. Bullied no more: when and how DNA shoves proteins around. Q Rev. Biophys. 45, 257–299 (2012).
Baranello, L., Levens, D., Gupta, A. & Kouzine, F. The importance of being supercoiled: How DNA mechanics regulate dynamic processes. Biochim. Biophys. Acta BBA - Gene Regul. Mech. 1819, 632–638 (2012).
Kouzine, F. et al. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat. Struct. Mol. Biol. 20, 396–403 (2013).
Chong, S., Chen, C., Ge, H. & Xie, X. S. Mechanism of transcriptional bursting in bacteria. Cell 158, 314–326 (2014).
Adrian, M. et al. Direct visualization of supercoiled DNA molecules in solution. EMBO J. 9, 4551–4554 (1990).
Boles, T. C., White, J. H. & Cozzarelli, N. R. Structure of plectonemically supercoiled DNA. J. Mol. Biol. 213, 931–951 (1990).
Bednar, J. et al. The twist, writhe and overall shape of supercoiled DNA change during counterion-induced transition from a loosely to a tightly interwound superhelix. Possible implications for DNA structure in vivo. J. Mol. Biol. 235, 825–847 (1994).
Levene, S. D., Donahue, C., Boles, T. C. & Cozzarelli, N. R. Analysis of the structure of dimeric DNA catenanes by electron microscopy. Biophys. J. 69, 1036–1045 (1995).
Cherny, D. I. & Jovin, T. M. Electron and scanning force microscopy studies of alterations in supercoiled DNA tertiary structure. J. Mol. Biol. 313, 295–307 (2001).
Amzallag, A. et al. 3D reconstruction and comparison of shapes of DNA minicircles observed by cryo-electron microscopy. Nucleic Acids Res. 34, e125 (2006).
Demurtas, D. et al. Bending modes of DNA directly addressed by cryo-electron microscopy of DNA minicircles. Nucleic Acids Res. 37, 2882–2893 (2009).
Lionberger, T. A. et al. Cooperative kinking at distant sites in mechanically stressed DNA. Nucleic Acids Res. 39, 9820–9832 (2011).
Fogg, J. M. et al. Exploring writhe in supercoiled minicircle DNA. J. Phys. Condens. Matter 18, S145–S159 (2006).
Cloutier, T. E. & Widom, J. Spontaneous sharp bending of double-stranded DNA. Mol. Cell 14, 355–362 (2004).
Cloutier, T. E. & Widom, J. DNA twisting flexibility and the formation of sharply looped protein-DNA complexes. Proc. Natl Acad. Sci. USA. 102, 3645–3650 (2005).
Bond, L. M., Peters, J. P., Becker, N. A., Kahn, J. D. & Maher, L. J. Gene repression by minimal lac loops in vivo. Nucleic Acids Res. 38, 8072–8082 (2010).
Shibata, Y. et al. Extrachromosomal microDNAs and chromosomal microdeletions in normal tissues. Science 336, 82–86 (2012).
Dubochet, J. et al. Cryo-electron microscopy of vitrified specimens. Q Rev. Biophys. 21, 129–228 (1988).
Depew, D. E. & Wang, J. C. Conformational fluctuations of DNA helix. Proc. Natl Acad. Sci. USA 72, 4275–4279 (1975).
Peters, J. P. & Maher, L. J. DNA curvature and flexibility in vitro and in vivo. Q Rev. Biophys. 43, 23–63 (2010).
Vetcher, A. A., McEwen, A. E., Abujarour, R., Hanke, A. & Levene, S. D. Gel mobilities of linking-number topoisomers and their dependence on DNA helical repeat and elasticity. Biophys. Chem. 148, 104–111 (2010).
Lau, P. P. & Gray, H. B. Extracellular nucleases of Alteromonas espejiana BAL 31.IV. The single strand-specific deoxyriboendonuclease activity as a probe for regions of altered secondary structure in negatively and positively supercoiled closed circular DNA. Nucleic Acids Res. 6, 331–357 (1979).
Kilpatrick, M. W., Wei, C. F., Gray, H. B. & Wells, R. D. BAL 31 nuclease as a probe in concentrated salt for the B-Z DNA junction. Nucleic Acids Res. 11, 3811–3822 (1983).
Allemand, J. F., Bensimon, D., Lavery, R. & Croquette, V. Stretched and overwound DNA forms a Pauling-like structure with exposed bases. Proc. Natl Acad. Sci. USA 95, 14152–14157 (1998).
Lankas, F., Lavery, R. & Maddocks, J. H. Kinking occurs during molecular dynamics simulations of small DNA minicircles. Structure. 14, 1527–1534 (2006).
Mitchell, J. S., Laughton, C. A. & Harris, S. A. Atomistic simulations reveal bubbles, kinks and wrinkles in supercoiled DNA. Nucleic Acids Res. 39, 3928–3938 (2011).
Randall, G. L., Zechiedrich, L. & Pettitt, B. M. In the absence of writhe, DNA relieves torsional stress with localized, sequence-dependent structural failure to preserve B-form. Nucleic Acids Res. 37, 5568–5577 (2009).
Du, Q., Kotlyar, A. & Vologodskii, A. Kinking the double helix by bending deformation. Nucleic Acids Res. 36, 1120–1128 (2008).
Benham, C. J. Energetics of the strand separation transition in superhelical DNA. J. Mol. Biol. 225, 835–847 (1992).
Benham, C. J. & Bi, C. The analysis of stress-induced duplex destabilization in long genomic DNA sequences. J. Comput. Biol. 11, 519–543 (2004).
Bi, C. & Benham, C. J. WebSIDD: server for predicting stress-induced duplex destabilized (SIDD) sites in superhelical DNA. Bioinforma. Oxf. Engl. 20, 1477–1479 (2004).
Pavlicek, J. W. et al. Supercoiling-induced DNA bending. Biochemistry 43, 10664–10668 (2004).
Ross, W. & Landy, A. Anomalous electrophoretic mobility of restriction fragments containing the att region. J. Mol. Biol. 156, 523–529 (1982).
Landy, A. Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu. Rev. Biochem. 58, 913–949 (1989).
Laundon, C. H. & Griffith, J. D. Curved helix segments can uniquely orient the topology of supertwisted DNA. Cell 52, 545–549 (1988).
Bliska, J. B. & Cozzarelli, N. R. Use of site-specific recombination as a probe of DNA structure and metabolism in vivo. J. Mol. Biol. 194, 205–218 (1987).
Hildebrandt, E. R. & Cozzarelli, N. R. Comparison of recombination in vitro and in E. coli cells: measure of the effective concentration of DNA in vivo. Cell 81, 331–340 (1995).
Kramer, P. R. & Sinden, R. R. Measurement of unrestrained negative supercoiling and topological domain size in living human cells. Biochemistry 36, 3151–3158 (1997).
Levchenko, V., Jackson, B. & Jackson, V. Histone release during transcription: displacement of the two H2A-H2B dimers in the nucleosome is dependent on different levels of transcription-induced positive stress. Biochemistry 44, 5357–5372 (2005).
Kouzine, F., Sanford, S., Elisha-Feil, Z. & Levens, D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat. Struct. Mol. Biol. 15, 146–154 (2008).
Dai, X., Greizerstein, M. B., Nadas-Chinni, K. & Rothman-Denes, L. B. Supercoil-induced extrusion of a regulatory DNA hairpin. Proc. Natl Acad. Sci. USA 94, 2174–2179 (1997).
Sandman, K., Grayling, R. A., Dobrinski, B., Lurz, R. & Reeve, J. N. Growth-phase-dependent synthesis of histones in the archaeon Methanothermus fervidus. Proc. Natl Acad. Sci. USA 91, 12624–12628 (1994).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Kremer, J. R., Mastronarde, D. N. & McIntosh, J. R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996).
Pettersen, E. F. et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Case, D. A. et al. AMBER 11 University of California (2010).
Pérez, A. et al. Refinement of the AMBER force field for nucleic acids: improving the description of alpha/gamma conformers. Biophys. J. 92, 3817–3829 (2007).
Krepl, M. et al. Reference simulations of noncanonical nucleic acids with different χ variants of the AMBER force field: quadruplex DNA, quadruplex RNA and Z-DNA. J. Chem. Theory Comput. 8, 2506–2520 (2012).
Harris, S. A., Laughton, C. A. & Liverpool, T. B. Mapping the phase diagram of the writhe of DNA nanocircles using atomistic molecular dynamics simulations. Nucleic Acids Res. 36, 21–29 (2008).
Tsui, V. & Case, D. A. Theory and applications of the generalized Born solvation model in macromolecular simulations. Biopolymers 56, 275–291 (2000).
Rybenkov, V. V., Vologodskii, A. V. & Cozzarelli, N. R. The effect of ionic conditions on DNA helical repeat, effective diameter and free energy of supercoiling. Nucleic Acids Res. 25, 1412–1418 (1997).
Shao, J., Tanner, S. W., Thompson, N. & Cheatham, T. E. Clustering Molecular Dynamics Trajectories: 1. Characterizing the Performance of Different Clustering Algorithms. J. Chem. Theory Comput. 3, 2312–2334 (2007).
Annapureddy, H. V. R. & Dang, L. X. Understanding the Rates and Molecular Mechanism of Water-Exchange around Aqueous Ions Using Molecular Simulations. J. Phys. Chem. B 118, 8917–8927 (2014).
Price, D. J. & Brooks, C. L. A modified TIP3P water potential for simulation with Ewald summation. J. Chem. Phys. 121, 10096–10103 (2004).
Pronk, S. et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinforma. Oxf. Engl. 29, 845–854 (2013).
Acknowledgements
We thank Dr Nancy Crisona (University of California, Berkeley) and Dr Kathleen Sandman (The Ohio State University) for the HMfB expression plasmid, pKS323, and invaluable advice on the expression, purification, and application of HMfB. Human topoisomerase IIα was kindly provided by Jo Ann Byl and Dr Neil Osheroff (Vanderbilt University School of Medicine). We thank Matthew Dougherty (Baylor College of Medicine) for help with generating Supplementary Movie 1. We thank Dr Richard Deibler for critically reading the manuscript and Prof. Nicola Stonehouse (University of Leeds) for advice on interpretation of the MD data. This research is partially supported by NIH grants P41GM103832 and P50GM1003297 to W.C., NIH grant R01GM080139 to S.J.L., NIH grants RO1A1054830 and R56AI054830 and the Human Frontier Science Program to L.Z., Biotechnology and Biological Sciences Research Council grant BB/I019472/1 to S.A.H. and NIH training grant (5T90DK070121-05) through the Gulf Coast Consortia to R.N.I.
Author information
Authors and Affiliations
Contributions
All electron microscopy was performed by R.N.I. Minicircle DNA samples were prepared by J.M.F. Analysis of cryo-ET data was performed by R.N.I., D.J.C, A.K.B., M.F.S. and L.Z. DNA relaxation experiments were performed by D.J.C. Gel electrophoresis and enzymatic and chemical probing for exposed bases were performed by J.M.F. Computational fitting of minicircle traces to cryo-ET densities was performed by M.C. and S.J.L. Molecular dynamics simulations were performed by T.S. and S.A.H. Figures were prepared by J.M.F., R.N.I., M.C. and T.S. Supplementary Figures were prepared by J.M.F, D.J.C. and T.S. All authors contributed to writing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
J.M.F., D.J.C., and L.Z. are co-inventors on several patents covering the minicircle technology and founders and shareholders in Twister Biotech. No other competing financial interests are reported.
Supplementary information
Supplementary Information
Supplementary Figures 1-5, Supplementary Tables 1-3, Supplementary Note 1 and Supplementary References (PDF 424 kb)
Supplementary Movie 1
An example of a tomogram obtained by cryo-ET. Dark densities represent volume rendering of the DNA minicircles. Red overlay was achieved by segmentation of the molecules. (MOV 12801 kb)
Supplementary Movie 2
Structure of an individual minicircle obtained by cryo-ET. The example shown adopts the handcuffs conformation. (MOV 4432 kb)
Supplementary Movie 3
Structure of an individual minicircle obtained by cryo-ET. The example shown adopts the needle conformation. (MOV 3959 kb)
Supplementary Movie 4
Implicit molecular dynamics simulation of ?Lk = -2. (MOV 12132 kb)
Supplementary Movie 5
Implicit molecular dynamics simulation of ?Lk = +1. (MOV 14698 kb)
Supplementary Movie 6
Explicit molecular dynamics simulation showing an open circle conformation. (MOV 3411 kb)
Supplementary Movie 7
Explicit molecular dynamics simulation showing a figure-8 conformation. (MOV 3405 kb)
Supplementary Movie 8
Explicit molecular dynamics simulations showing a racquet conformation. (MOV 3906 kb)
Supplementary Movie 9
Explicit molecular dynamics simulations showing a handcuffs conformation. (MOV 4569 kb)
Supplementary Movie 10
Atomistic MD structures from the explicitly solvated MD simulations embedded within the cryo-ET density maps showing the open circle conformation (Movie 10: RMSD = 26.4 Å, 8%). (The percentage value refers to the RMSD compared to the longest dimension of the tomogram). (MOV 22627 kb)
Supplementary Movie 11
Atomistic MD structures from the explicitly solvated MD simulations embedded within the cryo-ET density maps showing the figure-8 conformation (Movie 11: RMSD = 39.4 Å, 11%). (The percentage value refers to the RMSD compared to the longest dimension of the tomogram) (MOV 17073 kb)
Supplementary Movie 12
Atomistic MD structures from the explicitly solvated MD simulations embedded within the cryo-ET density maps showing the racquet conformation (Movie 12: RMSD = 35.5 Å, 10%). (The percentage value refers to the RMSD compared to the longest dimension of the tomogram) (MOV 11913 kb)
Supplementary Movie 13
Atomistic MD structures from the explicitly solvated MD simulations embedded within the cryo-ET density maps showing the handcuffs conformation (Movie 13: RMSD = 28.3 Å, 7%). (The percentage value refers to the RMSD compared to the longest dimension of the tomogram). (MOV 8874 kb)
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Irobalieva, R., Fogg, J., Catanese, D. et al. Structural diversity of supercoiled DNA. Nat Commun 6, 8440 (2015). https://doi.org/10.1038/ncomms9440
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/ncomms9440
This article is cited by
-
Angle between DNA linker and nucleosome core particle regulates array compaction revealed by individual-particle cryo-electron tomography
Nature Communications (2024)
-
Suitability of double-stranded DNA as a molecular standard for the validation of analytical ultracentrifugation instruments
European Biophysics Journal (2023)
-
Supercoiling and looping promote DNA base accessibility and coordination among distant sites
Nature Communications (2021)
-
WASP: a software package for correctly characterizing the topological development of ribbon structures
Scientific Reports (2021)
-
The biological applications of DNA nanomaterials: current challenges and future directions
Signal Transduction and Targeted Therapy (2021)