Abstract
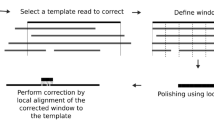
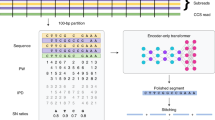
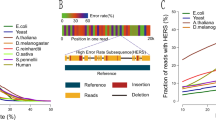
Single-molecule sequencing instruments can generate multikilobase sequences with the potential to greatly improve genome and transcriptome assembly. However, the error rates of single-molecule reads are high, which has limited their use thus far to resequencing bacteria. To address this limitation, we introduce a correction algorithm and assembly strategy that uses short, high-fidelity sequences to correct the error in single-molecule sequences. We demonstrate the utility of this approach on reads generated by a PacBio RS instrument from phage, prokaryotic and eukaryotic whole genomes, including the previously unsequenced genome of the parrot Melopsittacus undulatus, as well as for RNA-Seq reads of the corn (Zea mays) transcriptome. Our long-read correction achieves >99.9% base-call accuracy, leading to substantially better assemblies than current sequencing strategies: in the best example, the median contig size was quintupled relative to high-coverage, second-generation assemblies. Greater gains are predicted if read lengths continue to increase, including the prospect of single-contig bacterial chromosome assembly.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Margulies, M. et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380 (2005).
Bentley, D. Whole-genome re-sequencing. Curr. Opin. Genet. Dev. 16, 545–552 (2006).
Sanger, F., Nicklen, S. & Coulson, A. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74, 5463–5467 (1977).
Niu, B., Fu, L., Sun, S. & Li, W. Artificial and natural duplicates in pyrosequencing reads of metagenomic data. BMC Bioinformatics 11, 187 (2010).
Dohm, J., Lottaz, C., Borodina, T. & Himmelbauer, H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 36, e105 (2008).
Kingsford, C., Schatz, M. & Pop, M. Assembly complexity of prokaryotic genomes using short reads. BMC Bioinformatics 11, 21 (2010).
Schadt, E.E., Turner, S. & Kasarskis, A. A window into third-generation sequencing. Hum. Mol. Genet. 19, R227–R240 (2010).
Chin, C.-S. The origin of the Haitian cholera outbreak strain. N. Engl. J. Med. 364, 33–42 (2011).
Rasko, D.A. et al. Origins of the E. coli strain causing an outbreak of hemolytic–uremic syndrome in Germany. N. Engl. J. Med. 365, 709–717 (2011).
Eid, J. et al. Real-time DNA sequencing from single polymerase molecules. Science 323, 133–138 (2009).
Miller, J.R. et al. Aggressive assembly of pyrosequencing reads with mates. Bioinformatics 24, 2818–2824 (2008).
Salzberg, S.L. et al. GAGE: a critical evaluation of genome assemblies and assembly algorithms. Genome Res. 557–567 (2012).
Pop, M. Genome assembly reborn: recent computational challenges. Brief. Bioinform. 10, 354 (2009).
Miller, J., Koren, S. & Sutton, G. Assembly algorithms for next-generation sequencing data. Genomics 95, 315–327 (2010).
Phillippy, A., Schatz, M. & Pop, M. Genome assembly forensics: finding the elusive mis-assembly. Genome Biol. 9, R55 (2008).
Pevzner, P.A., Tang, H. & Waterman, M.S. An Eulerian path approach to DNA fragment assembly. Proc. Natl. Acad. Sci. USA 98, 9748–9753 (2001).
Schatz, M.C., Witkowski, J. & McCombie, W.R. Current challenges in de novo plant genome sequencing and assembly. Genome Biol. 13, 243 (2012).
Nagarajan, N. & Pop, M. Parametric complexity of sequence assembly: theory and applications to next generation sequencing. J. Comput. Biol. 16, 897–908 (2009).
Gnerre, S. et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl. Acad. Sci. USA 1513–1518 (2011).
Pop, M., Phillippy, A., Delcher, A.L. & Salzberg, S.L. Comparative genome assembly. Brief. Bioinform. 5, 237–248 (2004).
Schatz, M.C. et al. Hawkeye and AMOS: visualizing and assessing the quality of genome assemblies. Brief. Bioinform. published online, doi: 10.1093/bib/bbr074 (23 December 2011).
Sommer, D., Delcher, A., Salzberg, S. & Pop, M. Minimus: a fast, lightweight genome assembler. BMC Bioinformatics 8, 64 (2007).
Schatz, M.C., Delcher, A.L. & Salzberg, S.L. Assembly of large genomes using second-generation sequencing. Genome Res. 20, 1165–1173 (2010).
Earl, D.A. et al. Assemblathon 1: a competitive assessment of de novo short read assembly methods. Genome Res. 2224–2241 (2011).
Warren, W.C. et al. The genome of a songbird. Nature 464, 757–762 (2010).
Hillier, L. et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432, 695–716 (2004).
Vezzi, F., Narzisi, G. & Mishra, B. Feature-by-feature—evaluating de novo sequence assembly. PLoS ONE 7, e31002 (2012).
Wu, T.D. & Watanabe, C.K. Gmap: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21, 1859–1875 (2005).
Enard, W. et al. Molecular evolution of FOXP2, a gene involved in speech and language. Nature 418, 869–872 (2002).
Enard, W. FOXP2 and the role of cortico-basal ganglia circuits in speech and language evolution. Curr. Opin. Neurobiol. 21, 415–424 (2011).
Lai, C.S., Fisher, S.E., Hurst, J.A., Vargha-Khadem, F. & Monaco, A.P. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature 413, 519–523 (2001).
Haesler, S. et al. FoxP2 expression in avian vocal learners and non-learners. J. Neurosci. 24, 3164–3175 (2004).
Haesler, S. et al. Incomplete and inaccurate vocal imitation after knockdown of FoxP2 in songbird basal ganglia nucleus Area X. PLoS Biol. 5, e321 (2007).
Carroll, S.B. Evolution at two levels: on genes and form. PLoS Biol. 3, e245 (2005).
Brose, K. et al. Slit proteins bind Robo receptors and have an evolutionarily conserved role in repulsive axon guidance. Cell 96, 795–806 (1999).
Wada, K., Sakaguchi, H., Jarvis, E.D. & Hagiwara, M. Differential expression of glutamate receptors in avian neural pathways for learned vocalization. J. Comp. Neurol. 476, 44–64 (2004).
Maes, T., Barcelo, A. & Buesa, C. Neuron navigator: a human gene family with homology to unc-53, a cell guidance gene from Caenorhabditis elegans. Genomics 80, 21–30 (2002).
Matsunaga, E. & Okanoya, K. Vocal control area-related expression of neuropilin-1, plexin-A4, and the lig-and semaphorin-3A has implications for the evolution of the avian vocal system. Dev. Growth Differ. 51, 45–54 (2009).
Morgan, J.I. & Curran, T. Stimulus-transcription coupling in neurons: role of cellular immediate-early genes. Trends Neurosci. 12, 459–462 (1989).
Jarvis, E.D. & Nottebohm, F. Motor-driven gene expression. Proc. Natl. Acad. Sci. USA 94, 4097–4102 (1997).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Kent, W.J. Blat–the blast-like alignment tool. Genome Res. 12, 656–664 (2002).
Goldberg, S. et al. A Sanger/pyrosequencing hybrid approach for the generation of high-quality draft assemblies of marine microbial genomes. Proc. Natl. Acad. Sci. USA 103, 11240–11245 (2006).
Fraser, C.M., Eisen, J.A., Nelson, K.E., Paulsen, I.T. & Salzberg, S.L. The value of complete microbial genome sequencing (you get what you pay for). J. Bacteriol. 184, 6403–6405 (2002).
Li, Y. et al. Structural variation in two human genomes mapped at single-nucleotide resolution by whole genome de novo assembly. Nat. Biotechnol. 29, 723–730 (2011).
Feuk, L., Carson, A.R. & Scherer, S.W. Structural variation in the human genome. Nat. Rev. Genet. 7, 85–97 (2006).
Sebat, J. et al. Strong association of de novo copy number mutations with autism. Science 316, 445–449 (2007).
Rothberg, J.M. et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature 475, 348–352 (2011).
Li, R. et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 20, 265–272 (2010).
Kurtz, S. et al. Versatile and open software for comparing large genomes. Genome Biol. 5, R12 (2004).
Acknowledgements
We thank Pacific Biosciences, Roche 454, Illumina, BGI and the Duke Genome Center for the generation and/or release of many of the data sets examined herein, and to the Assemblathon working group for the coordination and release of the parrot genome data. This publication was developed and funded in part under Agreement No. HSHQDC-07-C-00020 awarded by the US Department of Homeland Security for the management and operation of the National Biodefense Analysis and Countermeasures Center (NBACC), a Federally Funded Research and Development Center. The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the US Department of Homeland Security. The Department of Homeland Security does not endorse any products or commercial services mentioned in this publication. The work conducted by the US Department of Energy Joint Genome Institute is supported by the Office of Science of the US Department of Energy under Contract No. DE-AC02-05CH11231. This work was also funded in part by the US National Institutes of Health (NIH) R01-HG006677-12 (M.C.S.), NIH 2R01GM077117-04A1 (B.P.W.), the state of Maryland (D.A.R.), National Science Foundation IOS-1032105 to W.R.M., and Howard Hughes Medical Institute and NIH Directors Pioneer Award to E.D.J.
Author information
Authors and Affiliations
Contributions
S.K. and A.M.P. conceived and designed the algorithm. S.K. implemented the algorithm and carried out the de novo assembly experiments. S.K., M.C.S. and A.M.P. drafted the manuscript, ran experiments and contributed analysis. B.P.W. modified the Celera Assembler to support long sequencing reads and developed the BOGART unitigger. J.M. and Z.W. sequenced Z. mays cDNA and performed analysis. J.H., G.G. and E.D.J. sequenced M. undulatus and performed analysis of vocal learning genes. D.A.R. provided and sequenced E. coli strains. W.R.M. sequenced S. cerevisiae S228c. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
W.R.M. has participated in Illumina-sponsored meetings over the past four years and received travel reimbursement and honoraria for presenting at these events.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1-7, Supplementary Notes 1–6 and Supplementary Figures 1–16 (PDF 2414 kb)
Supplementary Dataset 1
Celera Assembler and AMOS source code utilized for PacBio correction experiments described in this publication. (ZIP 9510 kb)
Supplementary Dataset 2
Celera Assembler source code utilized for PacBio assembly experiments described in this publication. (ZIP 3700 kb)
Rights and permissions
About this article
Cite this article
Koren, S., Schatz, M., Walenz, B. et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol 30, 693–700 (2012). https://doi.org/10.1038/nbt.2280
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.2280
This article is cited by
-
Conserved chromatin and repetitive patterns reveal slow genome evolution in frogs
Nature Communications (2024)
-
Complete genome analysis of pathogenic Metschnikowia bicuspidata strain MQ2101 isolated from diseased ridgetail white prawn, Exopalaemon carinicauda
BMC Microbiology (2023)
-
The Analysis of the Glycosyltransferase Gene Function From a Novel Granaticin Producer, Streptomyces Vilmorinianum. YP1
Current Microbiology (2023)
-
Mycoplasma agalactiae ST35: a new sequence type with a minimal accessory genome primarily affecting goats
BMC Veterinary Research (2022)
-
Genomic Analysis and Molecular Characteristics in Carbapenem-Resistant Klebsiella pneumoniae Strains
Current Microbiology (2022)