Abstract
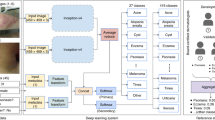
Skin cancer, the most common human malignancy1,2,3, is primarily diagnosed visually, beginning with an initial clinical screening and followed potentially by dermoscopic analysis, a biopsy and histopathological examination. Automated classification of skin lesions using images is a challenging task owing to the fine-grained variability in the appearance of skin lesions. Deep convolutional neural networks (CNNs)4,5 show potential for general and highly variable tasks across many fine-grained object categories6,7,8,9,10,11. Here we demonstrate classification of skin lesions using a single CNN, trained end-to-end from images directly, using only pixels and disease labels as inputs. We train a CNN using a dataset of 129,450 clinical images—two orders of magnitude larger than previous datasets12—consisting of 2,032 different diseases. We test its performance against 21 board-certified dermatologists on biopsy-proven clinical images with two critical binary classification use cases: keratinocyte carcinomas versus benign seborrheic keratoses; and malignant melanomas versus benign nevi. The first case represents the identification of the most common cancers, the second represents the identification of the deadliest skin cancer. The CNN achieves performance on par with all tested experts across both tasks, demonstrating an artificial intelligence capable of classifying skin cancer with a level of competence comparable to dermatologists. Outfitted with deep neural networks, mobile devices can potentially extend the reach of dermatologists outside of the clinic. It is projected that 6.3 billion smartphone subscriptions will exist by the year 2021 (ref. 13) and can therefore potentially provide low-cost universal access to vital diagnostic care.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
American Cancer Society. Cancer facts & figures 2016. Atlanta, American Cancer Society 2016. http://www.cancer.org/acs/groups/content/@research/documents/document/acspc-047079.pdf
Rogers, H. W. et al. Incidence estimate of nonmelanoma skin cancer (keratinocyte carcinomas) in the US population, 2012. JAMA Dermatology 151.10, 1081–1086 (2015)
Stern, R. S. Prevalence of a history of skin cancer in 2007: results of an incidence-based model. Arch. Dermatol. 146, 279–282 (2010)
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015)
LeCun, Y. & Bengio, Y. In The Handbook of Brain Theory and Neural Networks (ed. Arbib, M. A. ) 3361.10 (MIT Press, 1995)
Russakovsky, O. et al. Imagenet large scale visual recognition challenge. Int. J. Comput. Vis. 115, 211–252 (2015)
Krizhevsky, A., Sutskever, I. & Hinton, G. E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 25, 1097–1105 (2012)
Ioffe, S. & Szegedy, C. Batch normalization: accelerating deep network training by reducing internal covariate shift. Proc. 32nd Int. Conference on Machine Learning 448–456 (2015)
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J. & Wojna, Z. Rethinking the inception architecture for computer vision. Preprint at https://arxiv.org/abs/1512.00567 (2015)
Szegedy, C. et al. Going deeper with convolutions. Proc. IEEE Conference on Computer Vision and Pattern Recognition 1–9 (2015)
He, K. Zhang, X., Ren, S. & Sun J. Deep residual learning for image recognition. Preprint at https://arxiv.org/abs/1512.03385 (2015)
Masood, A. & Al-Jumaily, A. A. Computer aided diagnostic support system for skin cancer: a review of techniques and algorithms. Int. J. Biomed. Imaging 2013, 323268 (2013)
Cerwall, P. & Report, E. M. Ericssons mobility report https://www.ericsson.com/res/docs/2016/ericsson-mobility-report-2016.pdf (2016)
Rosado, B. et al. Accuracy of computer diagnosis of melanoma: a quantitative meta-analysis. Arch. Dermatol. 139, 361–367, discussion 366 (2003)
Burroni, M. et al. Melanoma computer-aided diagnosis: reliability and feasibility study. Clin. Cancer Res. 10, 1881–1886 (2004)
Kittler, H., Pehamberger, H., Wolff, K. & Binder, M. Diagnostic accuracy of dermoscopy. Lancet Oncol. 3, 159–165 (2002)
Codella, N. et al. In Machine Learning in Medical Imaging (eds Zhou, L., Wang, L., Wang, Q. & Shi, Y. ) 118–126 (Springer, 2015)
Gutman, D. et al. Skin lesion analysis toward melanoma detection. International Symposium on Biomedical Imaging (ISBI), (International Skin Imaging Collaboration (ISIC), 2016)
Binder, M. et al. Epiluminescence microscopy-based classification of pigmented skin lesions using computerized image analysis and an artificial neural network. Melanoma Res. 8, 261–266 (1998)
Menzies, S. W. et al. In Skin Cancer and UV Radiation (eds Altmeyer, P., Hoffmann, K. & Stücker, M. ) 1064–1070 (Springer, 1997)
Clark, W. H., et al. Model predicting survival in stage I melanoma based on tumor progression. J. Natl Cancer Inst. 81, 1893–1904 (1989)
Schindewolf, T. et al. Classification of melanocytic lesions with color and texture analysis using digital image processing. Anal. Quant. Cytol. Histol. 15, 1–11 (1993)
Ramlakhan, K. & Shang, Y. A mobile automated skin lesion classification system. 23rd IEEE International Conference on Tools with Artificial Intelligence (ICTAI) 138–141 (2011)
Ballerini, L. et al. In Color Medical Image Analysis. (eds, Celebi, M. E. & Schaefer, G. ) 63–86 (Springer, 2013)
Deng, J. et al. Imagenet: A large-scale hierarchical image database. EEE Conference on Computer Vision and Pattern Recognition 248–255 (CVPR, 2009)
Mnih, V. et al. Human-level control through deep reinforcement learning. Nature 518, 529–533 (2015)
Silver, D. et al. Mastering the game of Go with deep neural networks and tree search. Nature 529, 484–489 (2016)
Pan, S. J. & Yang, Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 22, 1345–1359 (2010)
Van der Maaten, L., & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008)
Abadi, M. et al. Tensorflow: large-scale machine learning on heterogeneous distributed systems. Preprint at https://arxiv.org/abs/1603.04467 (2016)
Acknowledgements
We thank the Thrun laboratory for their support and ideas. We thank members of the dermatology departments at Stanford University, University of Pennsylvania, Massachusetts General Hospital and University of Iowa for completing our tests. This study was supported by funding from the Baxter Foundation to H.M.B. In addition, this work was supported by a National Institutes of Health (NIH) National Center for Advancing Translational Science Clinical and Translational Science Award (UL1 TR001085). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author information
Authors and Affiliations
Contributions
A.E. and B.K. conceptualized and trained the algorithms and collected data. R.A.N., J.K. and S.S. developed the taxonomy, oversaw the medical tasks and recruited dermatologists. H.M.B. and S.T. supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information
Nature thanks A. Halpern, G. Merlino and M. Welling for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 Procedure for calculating inference class probabilities from training class probabilities.
Illustrative example of the inference procedure using a subset of the taxonomy and mock training/inference classes. Inference classes (for example, malignant and benign lesions) correspond to the red nodes in the tree. Training classes (for example, amelanotic melanoma, blue nevus), which were determined using the partitioning algorithm with maxClassSize = 1,000, correspond to the green nodes in the tree. White nodes represent either nodes that are contained in an ancestor node’s training class or nodes that are too large to be individual training classes. The equation represents the relationship between the probability of a parent node, u, and its children, C(u); the sum of the child probabilities equals the probability of the parent. The CNN outputs a distribution over the training nodes. To recover the probability of any inference node it therefore suffices to sum the probabilities of the training nodes that are its descendants. A numerical example is shown for the benign inference class: Pbenign = 0.6 = 0.1 + 0.05 + 0.05 + 0.3 + 0.02 + 0.03 + 0.05.
Extended Data Figure 2 Confusion matrix comparison between CNN and dermatologists.
Confusion matrices for the CNN and both dermatologists for the nine-way classification task of the second validation strategy reveal similarities in misclassification between human experts and the CNN. Element (i, j) of each confusion matrix represents the empirical probability of predicting class j given that the ground truth was class i, with i and j referencing classes from Extended Data Table 2d. Note that both the CNN and the dermatologists noticeably confuse benign and malignant melanocytic lesions—classes 7 and 8—with each other, with dermatologists erring on the side of predicting malignant. The distribution across column 6—inflammatory conditions—is pronounced in all three plots, demonstrating that many lesions are easily confused with this class. The distribution across row 2 in all three plots shows the difficulty of classifying malignant dermal tumours, which appear as little more than cutaneous nodules under the skin. The dermatologist matrices are each computed using the 180 images from the nine-way validation set. The CNN matrix is computed using a random sample of 684 images (equally distributed across the nine classes) from the validation set.
Extended Data Figure 3 Saliency maps for nine example images from the second validation strategy.
a–i, Saliency maps for example images from each of the nine clinical disease classes of the second validation strategy reveal the pixels that most influence a CNN’s prediction. Saliency maps show the pixel gradients with respect to the CNN’s loss function. Darker pixels represent those with more influence. We see clear correlation between the lesions themselves and the saliency maps. Conditions with a single lesion (a–f) tend to exhibit tight saliency maps centred around the lesion. Conditions with spreading lesions (g–i) exhibit saliency maps that similarly occupy multiple points of interest in the images. a, Malignant melanocytic lesion (source image: https://www.dermquest.com/imagelibrary/large/020114HB.JPG). b, Malignant epidermal lesion (source image: https://www.dermquest.com/imagelibrary/large/001883HB.JPG). c, Malignant dermal lesion (source image: https://www.dermquest.com/imagelibrary/large/019328HB.JPG). d, Benign melanocytic lesion (source image: https://www.dermquest.com/imagelibrary/large/010137HB.JPG). e, Benign epidermal lesion (source image: https://www.dermquest.com/imagelibrary/large/046347HB.JPG). f, Benign dermal lesion (source image: https://www.dermquest.com/imagelibrary/large/021553HB.JPG). g, Inflammatory condition (source image: https://www.dermquest.com/imagelibrary/large/030028HB.JPG). h, Genodermatosis (source image: https://www.dermquest.com/imagelibrary/large/030705VB.JPG). i, Cutaneous lymphoma (source image: https://www.dermquest.com/imagelibrary/large/030540VB.JPG).
Extended Data Figure 4 Extension of Figure 3 with a different dermatological question.
a, Identical plots and results as shown in Fig. 3a, except that dermatologists were asked if a lesion appeared to be malignant or benign. This is a somewhat unnatural question to ask, in the clinic, the only actionable decision is whether or not to biopsy or treat a lesion. The blue curves for the CNN are identical to Fig. 3. b, Figure 3b reprinted for visual comparison to a.
Rights and permissions
About this article
Cite this article
Esteva, A., Kuprel, B., Novoa, R. et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118 (2017). https://doi.org/10.1038/nature21056
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature21056