Abstract
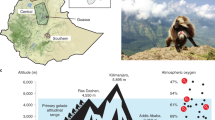
As modern humans migrated out of Africa, they encountered many new environmental conditions, including greater temperature extremes, different pathogens and higher altitudes. These diverse environments are likely to have acted as agents of natural selection and to have led to local adaptations. One of the most celebrated examples in humans is the adaptation of Tibetans to the hypoxic environment of the high-altitude Tibetan plateau1,2,3. A hypoxia pathway gene, EPAS1, was previously identified as having the most extreme signature of positive selection in Tibetans4,5,6,7,8,9,10, and was shown to be associated with differences in haemoglobin concentration at high altitude. Re-sequencing the region around EPAS1 in 40 Tibetan and 40 Han individuals, we find that this gene has a highly unusual haplotype structure that can only be convincingly explained by introgression of DNA from Denisovan or Denisovan-related individuals into humans. Scanning a larger set of worldwide populations, we find that the selected haplotype is only found in Denisovans and in Tibetans, and at very low frequency among Han Chinese. Furthermore, the length of the haplotype, and the fact that it is not found in any other populations, makes it unlikely that the haplotype sharing between Tibetans and Denisovans was caused by incomplete ancestral lineage sorting rather than introgression. Our findings illustrate that admixture with other hominin species has provided genetic variation that helped humans to adapt to new environments.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
13 August 2014
The affiliations list has been updated to correct the address of author Kui Li.
References
Moore, L. G., Young, D., McCullough, R. E., Droma, T. & Zamudio, S. Tibetan protection from intrauterine growth restriction (IUGR) and reproductive loss at high altitude. Am. J. Hum. Biol. 13, 635–644 (2001)
Niermeyer, S. et al. Child health and living at high altitude. Arch. Dis. Child. 94, 806–811 (2009)
Wu, T. et al. Hemoglobin levels in Quinghai-Tibet: different effects of gender for Tibetans vs. Han. J. Appl. Physiol. 98, 598–604 (2005)
Yi, X. et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329, 75–78 (2010)
Bigham, A. et al. Identifying signature of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 6, e1001116 (2010)
Simonson, T. S. et al. Genetic evidence for high-altitude adaptation in Tibet. Science 329, 72–75 (2010)
Beall, C. M. et al. Natural selection on EPAS1 (HIF2a) associated with low hemoglobin concentration in Tibetan highlanders. Proc. Natl Acad. Sci. USA 107, 11459–11464 (2010)
Peng, Y. et al. Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Mol. Biol. Evol. 28, 1075–1081 (2011)
Xu, S. et al. A genome-wide search for signals of high-altitude adaptation in Tibetans. Mol. Biol. Evol. 28, 1003–1011 (2011)
Wang, B. et al. On the origin of Tibetans and their genetic basis in adapting high-altitude environments. PLoS ONE 6, e17002 (2011)
Moore, L. G. et al. Maternal adaptation to high-altitude pregnancy: an experiment of nature—a review. Placenta 25, S60–S71 (2004)
Vargas, E. & Spielvogel, H. Chronic mountain sickness, optimal hemoglobin, and heart disease. High Alt. Med. Biol. 7, 138–149 (2006)
Yip, R. Significance of an abnormally low or high hemoglobin concentration during pregnancy: special consideration of iron nutrition1'2'3. Am. J. Clin. Nutr. 72, 272S–279S (2000)
Meyer, M. et al. A high-coverage genome sequence from an archaic Denisovan individual. Science 338, 222–226 (2012)
Li, J. Z. et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science 319, 1100–1104 (2008)
Rosenberg, N. A. Standardized subsets of the HGDP-CEPH Human Genome Diversity Cell Line Panel, accounting for atypical and duplicated samples and pairs of close relatives. Ann. Hum. Genet. 70, 841–847 (2006)
Soejima, M. & Koda, Y. Population differences of two coding SNPs. in pigmentation-related genes SLC24A5 and SLC45A2. Int. J. Legal Med. 121, 36–39 (2007)
Sulem, P. et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nature Genet. 39, 1443–1452 (2007)
Coop, G. et al. The role of geography in human adaptation. PLoS Genet. 5, e1000500 (2009)
Pickrell, J. K. et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 19, 826–837 (2009)
An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012)
Paradis, E. Pegas: an R package for population genetics with an integrated–modular approach. Bioinformatics 26, 419–420 (2010)
Vernot, B. & Akey, J. Resurrecting Surviving neandertal lineages from modern human genomes. Science (2014)
Plagnol, V. & Wall, J. D. Possible ancestral structure in human populations. PLoS Genet. 2, e105 (2006)
Reich, D. et al. Genetic history of an archaic hominin group from Denisova cave in Siberia. Nature 468, 1053–1060 (2010)
Prüfer, K. et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49 (2014)
Skoglund, P. & Jakobsson, M. Archaic human ancestry in East Asia. Proc. Natl Acad. Sci. USA 108, 18301–18306 (2011)
Abi-Rached, L. et al. The shaping of modern human immune systems by multiregional admixture with archaic humans. Science 334, 89–94 (2011)
Mendez, F. L., Watkins, J. C. & Hammer, M. F. A haplotype at STAT2 introgressed from Neanderthals and serves as a candidate of positive selection in Papua New Guinea. Am. J. Hum. Genet. 91, 265–274 (2012)
Sankararaman, S. et al. The genomic landscape of Neanderthal ancestry in present-day humans. Nature (2014)
Li, R., Li, Y., Kristiansen, K. & Wang, J. SOAP: short oligonucleotide alignment program. Bioinformatics 24, 713–714 (2008)
Li, R. et al. SNP detection for massively parallel whole-genome resequencing. Genome Res. 19, 1124–1132 (2009)
Browning, B. L. & Browning, S. R. A fast, powerful method for detecting identity by descent. Am. J. Hum. Genet. 88, 173–182 (2011)
Coop, G. et al. The role of geography in human adaptation. PLoS Genet. 5, e1000500 (2009)
Reynolds, J., Weir, B. S. & Cockerham, C. C. Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics 105, 767–779 (1983)
R Development Core Team R: A language and environment for statistical computing http://www.R-project.org/ (R Foundation for Statistical Computing, 2011)
Ewing, G. & Hermisson, J. MSMS: a coalescent simulation program including recombination, demographic structure, and selection at a single locus. Bioinformatics 26, 2064–2065 (2010)
Myers, S. et al. A fine-scale map of recombination rates and hotspots across the human genome. Science 310, 321–324 (2005)
Hinch, A. G. et al. The landscape of recombination in African Americans. Nature 476, 170–175 (2011)
Scally, A. & Durbin, R. Revising the human mutation rate: implications for understanding human evolution. Nature Rev. Genet. 13, 745–753 (2012)
Teshima, K. M. & Innan, H. mbs: modifying Hudson’s ms software to generate samples of DNA sequences with a biallelic site under selection. BMC Bioinformatics 10, 166 (2009)
Hudson, R. R. Generating samples under a Wright–Fisher neutral model of genetic variation. Bioinformatics 18, 337–338 (2002)
Sankararaman, S. et al. The date of interbreeding between Neandertals and modern humans. PLoS Genet. 8, e1002947 (2012)
Durand, E. Y. et al. Testing for ancient admixture between closely related populations. Mol. Biol. Evol. 28, 2239–2252 (2011)
Simonson, T. S. et al. Genetic evidence for high-altitude adaptation in Tibet. Science 329, 72–75 (2010)
Acknowledgements
This research was funded by the State Key Development Program for Basic Research of China, 973 Program (2011CB809203, 2012CB518201, 2011CB809201, 2011CB809202), China National GeneBank-Shenzhen and Shenzhen Key Laboratory of Transomics Biotechnologies (no. CXB201108250096A). This work was also supported by research grants from the US NIH; R01HG003229 to R.N. and R01HG003229-08S2 to E.H.S. We thank F. Jay, M. Liang and F. Casey for useful discussions.
Author information
Authors and Affiliations
Contributions
R.N., Ji.W. and Ju.W. supervised the project. X.J., A., Z.B., Y.L., X.Y., M.H., P.N., B.W., X.O., H., J.L., Z.X.P.C., K.L., G.G., Y.Y., W.W., X.Z., X.X., H.Y., Y.L., Ji.W. and Ju.W. collected and generated the data, and performed the preliminary bioinformatic analyses to call SNPs and indels from the raw data. E.H.-S. and N.V. filtered the data and B.M.P. phased the data. E.H.-S. performed the majority of the population genetic analysis with some contributions from B.M.P. and M.S. E.H.-S. and R.N. wrote the manuscript with critical input from all the authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
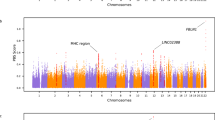
Extended Data Figure 1 FST calculated for each SNP between Tibetan and Han populations.
Each dot represents the FST value for each SNP in EPAS1. The x axis is the physical position in the gene. Positions are based on the hg18 build of the human genome. The green box defines a 32.7-kb region where we observe the largest genetic differentiation between Han Chinese and Tibetans. The first and last positions of this 32.7-kb region correspond to the first and last position of the SNPs listed in Supplementary Table 3. For comparison, in ref. 4 the genome-wide FST between Han and Tibetans is 0.02. The site with the largest frequency difference (and therefore largest FST) is circled.
Extended Data Figure 2 Distribution of fixed differences.
The left panel is the distribution of fixed differences between two haplotype groups under a scenario of selection on a de novo mutation (see Methods), and the right panel is the distribution under a scenario of selection on standing variation (see Methods) for a region of size ∼32.7 kb. The initial frequency of the selected allele in the SSV model is 1%. Each row of panels corresponds to different selection strengths (2Ns) from 200 to 1,000. The red lines mark the number of fixed differences observed between the two haplotype classes in the real data for the given window size.
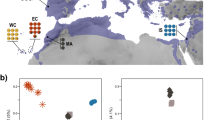
Extended Data Figure 3 Haplotype frequencies for Tibetans, our Han samples and the populations from the 1000 genomes project for the five-SNP motif in the EPAS1 region.
The y axis is the haplotype frequency. The legend shows all the possible haplotypes for the region considered among these populations: ASW, African American from the south western United States; CEU, Utah Residents with Northern and Western European ancestry; CHB, Han Chinese from Beijing; CHS, Southern Han Chinese; CLM, Colombian; FIN, Finnish; GBR, British; HAN, Han Chinese from Beijing; IBS, Iberian; JPT, Japanese; MXL, Mexican; PUR, Puerto Rican; LWK, Luhya; TSI, Toscani; TIB, Tibetan; YRI, Yoruban (see Methods).
Extended Data Figure 4 Derived allele frequency of the SNPs with the largest frequency difference between Tibetans and the 1000 Genomes Project populations.
At these SNPs, the frequency difference between Tibetans and the 1000 Genomes project populations is 0.65 or larger. Positions 46571435, 46579689, 46584859 and 46600358 were not called as SNPs in the 1000 Genomes data, so we assume these positions were fixed for the human reference allele. Note that even though position 46577251, 46588331, 46594122 and 46598025 appear to have a frequency of 0.0 for the populations in the 1000 Genomes data, the derived allele in these SNPs are observed at very low frequency in at least one population (for example, CHB).
Extended Data Figure 5 Differences between haplotypes.
a, The full matrix of pairwise differences between all the unique haplotypes in Fig. 3, for the 40 most common haplotypes identified in the 1000 Genomes and the Tibetan samples in the 32.7-kb region of EPAS1. The Denisovan haplotype (of frequency two) was added afterwards for comparison. The unique haplotypes are labelled with Roman numerals (here and in Fig. 3), and the Denisovan haplotype is the first column, haplotype I. Refer to Fig. 3 in the main text and the supplementary material for the representation of populations for each haplotype. b, Illustration of the genealogical structure in a model with gene flow from Denisovans to Tibet. Letters a–k are the labels for the branch lengths and are adjacent to their corresponding branches. The divergence between modern human haplotypes and the introgressed haplotype in Tibetans would be larger than the haplotypes in other modern human populations and the Denisovan haplotype (see Methods and Supplementary Information). TIB, CEU and YRI denote Tibetan, European and Yoruban populations. Note that the lengths i and k are unknown as we do not know when these populations went extinct.
Extended Data Figure 6 Other haplotype networks.
a, A haplotype network based on the number of pairwise differences between 43 unique haplotypes defined from the 20 most differentiated SNPs between Tibetans and the 14 populations from the 1000 Genomes Project. The R software package pegas (ref. 22) was used to generate the figure. The haplotype distances are from pairwise differences. Each pie chart represents one unique haplotype and the size of the pie chart is proportional to log2(number of chromosomes with that haplotype). The sections in the pie provide the breakdown of the haplotypes amongst populations. The width of the edges is proportional to the number of pairwise differences between the joined haplotypes; the thinnest edge width represents a difference of one mutation. The number 57 next to a Tibetan haplotype is the number of Tibetan chromosomes with that haplotype. Similarly, the number 1,912 is the number of chromosomes (across several populations) with that haplotype. b, The number of pairwise differences between the Denisovan haplotype and the 43 unique haplotypes defined from the 20 most differentiated SNPs between Tibetans and the 14 populations from the 1000 Genomes Project (same haplotypes as in a). Each bar is a unique haplotype, and they are sorted in increasing order of pairwise differences. The colours within each bar represent the proportion of chromosomes with that haplotype broken down by populations. The numbers on top of each bar represent the total number of chromosomes within the 1000 Genomes data set and Tibetans that have the haplotype. Note this is the same data set used to create the haplotype network in panel a. Supplementary Tables 5 and 6 contain the 43 haplotypes and the frequencies within each of the populations.
Extended Data Figure 7 Number of pairwise differences.
Red bars are the histograms of the number of pairwise differences between Denisovan and Tibetans. Blue bars are the histograms of the number of pairwise differences between Denisovan and GBR, CHS, FIN, PUR, CLM, IBS, CEU, YRI, CHB, JPT, LWK, ASW, MXL or TSI. All comparisons are within the 32.7-kb region of high differentiation (green box in Extended Data Fig. 1).
Extended Data Figure 8 Divergence distributions.
Modern human–Denisovan divergence (see Methods) for intronic regions of size 32.7 kb is plotted in red. Modern human–modern human divergence for the same intronic regions is plotted in blue. At the EPAS1 32.7-kb region, in green, is plotted the Tibetan–Han divergence. The black arrow points to the number of nucleotide differences between the Denisovan and the most common Tibetan haplotype (0.0038). This value is significantly lower than what we observe between modern human–Denisovan (red curve, P = 0.0028).
Extended Data Figure 9 Null distributions of D for an assumed Tibet–Han divergence of 3,000 years.
Each histogram corresponds to the D values obtained under null models without gene flow, and the red vertical bar corresponds to the D values observed in the real data. The observed D values are significant (P < 0.001) even when we assume Tibet–Han divergence of 5,000 or 10,000 years (see Methods and Supplementary Tables 8–10) (model abbreviations are given in the Supplementary Information; section on D statistics under models of no gene flow).
Extended Data Figure 10 S* statistics and PCA plot.
a, A measure of introgression, S*, from ref. 23. Distributions are for 1,000 simulations under the four demographic models described in the Supplementary Information; section on D statistics under models of no gene flow. S* for the Tibetan individuals is shown as a vertical grey line. For all models, the empirical P values are 0.035, 0.028, 0.019 and 0.017, respectively, for each model (top to bottom). b, Plots the first and second principal components using all the CHS (100 individuals) and the CHB (97 individuals) from the 1000 Genomes and the 77 Tibetan individuals from ref. 45 (see Methods). The black circle and the black triangle represent the single CHB and the CHS individuals carrying the five-SNP Tibetan–Denisovan-haplotype (Extended Data Fig. 3). All SNPs in the intersection between the 1000 Genomes populations and the 77 Tibetan individuals from chromosome 2 were used for this analysis.
Supplementary information
Supplementary Information
This file contains Supplementary Text, Supplementary References and Supplementary Tables 1-11. (PDF 342 kb)
Rights and permissions
About this article
Cite this article
Huerta-Sánchez, E., Jin, X., Asan et al. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature 512, 194–197 (2014). https://doi.org/10.1038/nature13408
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13408
This article is cited by
-
Differentiated adaptative genetic architecture and language-related demographical history in South China inferred from 619 genomes from 56 populations
BMC Biology (2024)
-
Ancient and recent origins of shared polymorphisms in yeast
Nature Ecology & Evolution (2024)
-
New dating indicates intermittent human occupation of the Nwya Devu Paleolithic site on the high-altitude central Tibetan Plateau during the past 45,000 years
Science China Earth Sciences (2024)
-
North African fox genomes show signatures of repeated introgression and adaptation to life in deserts
Nature Ecology & Evolution (2023)
-
Humans have lived on the Tibetan Plateau for 5,000 years
Nature (2023)