Abstract
The silkworm Bombyx mori uses a WZ sex determination system that is analogous to the one found in birds and some reptiles. In this system, males have two Z sex chromosomes, whereas females have Z and W sex chromosomes. The silkworm W chromosome has a dominant role in female determination1,2, suggesting the existence of a dominant feminizing gene in this chromosome. However, the W chromosome is almost fully occupied by transposable element sequences3,4,5, and no functional protein-coding gene has been identified so far. Female-enriched PIWI-interacting RNAs (piRNAs) are the only known transcripts that are produced from the sex-determining region of the W chromosome6, but the function(s) of these piRNAs are unknown. Here we show that a W-chromosome-derived, female-specific piRNA is the feminizing factor of B. mori. This piRNA is produced from a piRNA precursor which we named Fem. Fem sequences were arranged in tandem in the sex-determining region of the W chromosome. Inhibition of Fem-derived piRNA-mediated signalling in female embryos led to the production of the male-specific splice variants of B. mori doublesex (Bmdsx), a gene which acts at the downstream end of the sex differentiation cascade7,8. A target gene of Fem-derived piRNA was identified on the Z chromosome of B. mori. This gene, which we named Masc, encoded a CCCH-type zinc finger protein. We show that the silencing of Masc messenger RNA by Fem piRNA is required for the production of female-specific isoforms of Bmdsx in female embryos, and that Masc protein controls both dosage compensation and masculinization in male embryos. Our study characterizes a single small RNA that is responsible for primary sex determination in the WZ sex determination system.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
GenBank/EMBL/DDBJ
Data deposits
The nucleotide sequences of Fem and Masc have been deposited in the DDBJ/EMBL/GenBank data bank under the accession numbers AB840787 and AB840788. Deep sequencing data obtained in this study are available under the accession numbers DRA001104 and DRA001338 (DDBJ), respectively.
References
Hasimoto, H. The role of the W-chromosome in the sex determination of Bombyx mori [in Japanese]. Jpn. J. Genet. 8, 245–247 (1933)
Tajima, Y. Studies on chromosome aberrations in the silkworm. II. Translocation involving second and W-chromosomes [in Japanese]. Bull. Seric. Exp. Stn. 12, 109–181 (1944)
Abe, H. et al. Identification of novel random amplified polymorphic DNAs (RAPDs) on the W chromosome of the domesticated silkworm, Bombyx mori, and the wild silkworm, B. mandarina, and their retrotransposable element-related nucleotide sequences. Genes Genet. Syst. 73, 243–254 (1998)
Abe, H. et al. Partial deletions of the W chromosome due to reciprocal translocation in the silkworm Bombyx mori. Insect Mol. Biol. 14, 339–352 (2005)
Abe, H., Mita, K., Yasukochi, Y., Ohshiki, T. & Shimada, T. Retrotransposable elements on the W chromosome of the silkworm, Bombyx mori. Cytogenet. Genome Res. 110, 144–151 (2005)
Kawaoka, S. et al. The silkworm W chromosome is a source of female-enriched piRNAs. RNA 17, 2144–2151 (2011)
Suzuki, M. G., Funaguma, S., Kanda, T., Tamura, T. & Shimada, T. Analysis of the biological functions of a doublesex homologue in Bombyx mori. Dev. Genes Evol. 213, 345–354 (2003)
Suzuki, M. G., Funaguma, S., Kanda, T., Tamura, T. & Shimada, T. Role of the male BmDSX protein in the sexual differentiation of Bombyx mori. Evol. Dev. 7, 58–68 (2005)
Ohbayashi, F., Suzuki, M. G., Mita, K., Okano, K. & Shimada, T. A homologue of the Drosophila doublesex gene is transcribed into sex-specific mRNA isoforms in the silkworm, Bombyx mori. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 128, 145–158 (2001)
The International Silkworm Genome Consortium. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 38, 1036–1045 (2008)
Kawaoka, S. et al. Bombyx small RNAs: genomic defense system against transposons in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 38, 1058–1065 (2008)
Kawaoka, S. et al. Zygotic amplification of secondary piRNAs during silkworm embryogenesis. RNA 17, 1401–1407 (2011)
Hara, K. et al. Altered expression of testis-specific genes, piRNAs, and transposons in the silkworm ovary masculinized by a W chromosome mutation. BMC Genomics 13, 119 (2012)
Kawaoka, S. et al. The Bombyx ovary-derived cell line endogenously expresses PIWI/PIWI-interacting RNA complexes. RNA 15, 1258–1264 (2009)
Kawaoka, S., Minami, K., Katsuma, S., Mita, K. & Shimada, T. Developmentally synchronized expression of two Bombyx mori Piwi subfamily genes, SIWI and BmAGO3 in germ-line cells. Biochem. Biophys. Res. Commun. 367, 755–760 (2008)
Brennecke, J. et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103 (2007)
Gunawardane, L. S. et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315, 1587–1590 (2007)
Watanabe, T. et al. Role of piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science 332, 848–852 (2011)
Sugimoto, T. N. & Ishikawa, Y. A male-killing Wolbachia carries a feminizing factor and is associated with degradation of the sex-determining system of its host. Biol. Lett. 8, 412–415 (2012)
Penalva, L. O. & Sánchez, L. RNA binding protein sex-lethal (Sxl) and control of Drosophila sex determination and dosage compensation. Microbiol. Mol. Biol. Rev. 67, 343–359 (2003)
Traut, W., Sahara, K. & Marec, F. Sex chromosomes and sex determination in Lepidoptera. Sex Dev. 1, 332–346 (2007)
Sakai, H., Yokoyama, T., Abe, H., Fujii, T. & Suzuki, M. G. Appearance of differentiated cells derived from polar body nuclei in the silkworm, Bombyx mori. Front. Physiol. 4, 235 (2013)
Sato, Y. et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nature Genet. 45, 860–867 (2013)
Sakudoh, T. et al. Diversity in copy number and structure of a silkworm morphogenetic gene as a result of domestication. Genetics 187, 965–976 (2011)
Suzuki, M. G. et al. Establishment of a novel in vivo sex-specific splicing assay system to identify a trans-acting factor that negatively regulates splicing of Bombyx mori dsx female exons. Mol. Cell. Biol. 28, 333–343 (2008)
Wang, L. et al. Mutation of a novel ABC transporter gene is responsible for the failure to incorporate uric acid in the epidermis of ok mutants of the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 43, 562–571 (2013)
Yamaguchi, J., Mizoguchi, T. & Fujiwara, H. siRNAs induce efficient RNAi response in Bombyx mori embryos. PLoS ONE 6, e25469 (2011)
Hutvágner, G., Simard, M. J., Mello, C. C. & Zamore, P. D. Sequence-specific inhibition of small RNA function. PLoS Biol. 2, e98 (2004)
Shoji, K. et al. Characterization of a novel chromodomain-containing gene from the silkworm, Bombyx mori. Gene 527, 649–654 (2013)
Katsuma, S. et al. Novel macula-like virus identified in Bombyx mori cultured cells. J. Virol. 79, 5577–5584 (2005)
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnol. 29, 644–652 (2011)
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011)
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010)
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009)
Abe, H. et al. Identification of the female-determining region of the W chromosome in Bombyx mori. Genetica 133, 269–282 (2008)
Fujii, T. et al. The female killing chromosome of the silkworm, Bombyx mori, was generated by translocation between the Z and W chromosomes. Genetica 127, 253–265 (2006)
Reuter, M. et al. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480, 264–267 (2011)
Acknowledgements
We thank S. G. Kamita for critical reading of the manuscript; Y. Tomari for critical reading of the manuscript and technical suggestions. This work was supported by the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry to Su.K. and Grants-in-Aid for Scientific Research on Innovative Areas (Nos. 22115502 and 22128004) to Su.K. and T.S.
Author information
Authors and Affiliations
Contributions
Su.K., T.K. and M.G.S. conceived and designed the experiments. T.K., H.K., K.S., H.S., G.I., Y.A., Sh.K., M.G.S. and Su.K. performed molecular biological experiments. M.K. and K.S. performed most of the bioinformatic analyses. S.S. and Y.S. performed deep sequencing and data analysis. T.S. provided essential reagents and expertise. All of the authors discussed the data and helped manuscript preparation. Su.K. wrote the manuscript with intellectual input from all authors. Su.K. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
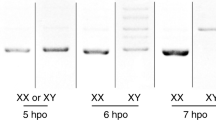
Extended Data Figure 1 Molecular sexing and comparative transcriptome analysis of embryonic B. mori.
a, Molecular sexing of individual embryos at 21 hpo. Musashi, Sasuke and Bonsai are W chromosome RAPD markers. ‘Chr2’ control bands are generated from a primer set that amplifies a sequence within the 2nd chromosome of B. mori. b, MA plots of RNA-seq data. The comp73859_c0 contig is indicated by red dots and highlighted by arrows. The axes show: A (x-axis) = (log2(transcripts per million in male) + log2(transcripts per million in female))/2. M (y-axis) = log2(transcripts per million in male) − log2(transcripts per million in female). c, Number of the comp73859_c0-derived transcripts in each RNA-seq library. Note that the comp73859_c0-derived transcripts detected in male libraries may be derived from incorrectly sexed embryos or RNA produced by polar bodies. Combined with RT–qPCR results of Fig. 1c, the expression level of this contig peaks around 18–21 hpo in the B. mori embryo.
Extended Data Figure 2 Expression profile of the female-specific comp73859_c0 contig.
a, Developmental expression profile of the female-specific contig in ovary during the larval (4th and 5th instars) and pupal stages. RT–qPCR was performed using total RNA that was isolated from ovary of 4th and 5th instar larvae, and pupae (p50T). This contig was detected in the ovary of 4th and 5th instar larvae, and pupae of B. mori with a strong peak expression at an early pupal stage. rp49 was used as an internal control. Data shown are mean + s.d. of three individuals, except for day 0 of 5th instar (n = 2). b, The mRNA expression in 17 different tissues from day 3, 5th instar larvae (p50T). RT–qPCR was performed using total RNA from brain (BR), prothoracic gland (PG), salivary gland (SG), fat body (FB), trachea (TR), haemocyte (HC), testis (TES), ovary (OV), anterior silkgland (ASG), middle silkgland (MSG), posterior silkgland (PSG), foregut (FG), midgut (MG), hindgut (HG), Malpighian tubules (MT), integument (IG) of male and female larvae (except for testis and ovary) or BmN4 cells (BmN). rp49 was used as an internal control. c, Amplification of the female-specific transcript. Long PCR using female gDNA and cDNA as templates was performed with primers 1F and 1R. Black arrows show bands corresponding to single or multiple units of this transcript. The predicted structure of each unit was also indicated. d, Northern blot analysis of total RNA that was prepared from embryos (24 hpo) and tissues from day 3 5th instar F1 hybrid Kinshu × Showa larvae (ovary, testis, fat body, and silk gland), and BmN cells. The asterisks show major transcripts.
Extended Data Figure 3 Characterization of the female-specific piRNA.
a, Detection of contig-derived piRNA and piRNA-1 (control). Northern blot analysis was performed using total RNA prepared from early embryos. The asterisks show the location of each piRNA. b, Normalized reads of the female-specific piRNA in embryonic piRNA libraries of B. mori12 generated at 0, 6, 12, and 24 hpo. Reads of 26–29 nucleotides that showed 2 or fewer mismatches to the corresponding piRNA sequence were scored as a positive match. c, RT–qPCR estimation of the female-specific piRNA levels in early embryos. The piRNA level was normalized to that of let-7. d, Normalized reads of the female-specific piRNA in piRNA libraries6 from ovary and testis of wild-type B. mori or W chromosome mutants. Schematic representation of sex chromosomes of each strain is shown below the panel. The putative sex-determining region is represented by the green box. The orange bar represents the W chromosome derived from B. mandarina. OV, ovary from wild-type; TES, testis from wild-type; MW, ovary from MW strain; LY, ovary from LY strain; WF, testis from WF strain.
Extended Data Figure 4 Effect of inhibition of the piRNA pathway on the splicing of Bmdsx transcripts.
a, Abundance of the female-specific piRNA in three piRNA libraries constructed from three KG individual ovaries (KG12, KG41 and KG42)13. Of these, two (KG41 and KG42) showed a severe masculinized phenotype, and the rest (KG12) showed a weak phenotype. KG12 expressed a slightly lower amount of this piRNA than that of LY (82.4% of LY), whereas its expression in the ovary of severe masculinized individuals (KG41 and KG42) were markedly lower than LY’s (12.1 and 29.7% of LY, respectively). The abbreviations are the same as in Extended Data Fig. 3d. b, Effect of the inhibitor RNA on the Bmdsx splicing. BmN4 cells were transfected with the inhibitor RNA or control RNA (that is, inhibitor for GFP piRNA), and the splicing patterns of Bmdsx were examined by RT–PCR. The F and M indicate female- and male-type splicing of Bmdsx, respectively. Similar results were obtained in three independent experiments. c, Effect of the RNA inhibitor on the Bmdsx splicing. The Bmdsx splicing patterns were examined at about 240 h post-injection (immediately after hatching). The abbreviations are the same as in Fig. 2c. The number indicates the sample size. d, Knockdown of Siwi or BmAgo3 mRNAs in female and male embryos. The embryos were injected with two types of siRNAs that target Siwi (Siwi-1 and Siwi-2) or BmAgo3 (Ago3-1 and Ago3-2) or a control siRNA that targets GFP. Total RNA was isolated from female or male siRNA-injected embryos at 72 h post-injection and RT–qPCR was performed. The data shown are mean + s.d. The number above each bar indicates the sample size of each group. e, Representative patterns of the Bmdsx splicing in siRNA-injected embryos. The F and M indicate female- and male-type splicing of Bmdsx, respectively.
Extended Data Figure 5 Characterization of Masc.
a, Structure of Masc mRNA. Five Masc transcripts (A–E) that encode full-length Masc proteins but show unique splicing patterns in the 3′-untranslated region as well as one transcript (F) that encodes a truncated Masc protein are found. b, Domain structure of the Masc protein. The hexagons show the location of two CCCH-type zinc finger domains. The amino acid sequences of these domains are shown below. The conserved CCCH residues are shown in red. c, Phylogenetic analysis of Masc proteins. The neighbour-joining tree was generated using the amino acid sequences of zinc finger domains from proteins showing homology to Masc. The numbers on the internal branches represent the support value in the bootstraps of 1,000 replicates.
Extended Data Figure 6 Cleavage of Masc mRNA.
a, Identification of the cleavage site of Masc mRNA. The Masc mRNA-derived RNA fragments were amplified by a modified RACE method, cloned, and sequenced. The RACE adaptor and the cloned 5′-end are indicated. Thirteen 5′-ends were determined and all showed identical sequences. Nucleotides identical to the top sequence are represented by asterisks. b, Detection of Masc piRNA. Northern blot analysis was performed using total RNA prepared from early embryos. The asterisk shows the location of Masc piRNA. c, Mapping of embryonic piRNAs (24 hpo) onto Masc mRNA. The relative location of ORF of Masc is shown below. d, Normalized reads of Fem piRNA and Masc piRNA in Siwi- or BmAgo3-immunoprecipitated libraries from BmN4 cells14. e, A ping-pong amplification model of Fem piRNA/Masc piRNA. f, Normalized reads of Masc piRNA in embryonic piRNA libraries. Reads of 26–29 nucleotides that showed 2 or fewer mismatches to the Masc piRNA sequence were scored as positive. g, RT–qPCR estimation of Masc piRNA in early embryos. The Masc piRNA level was normalized to that of let-7.
Extended Data Figure 7 Effects of Siwi or BmAgo3 knockdown on the Bmdsx splicing and Masc expression.
a, Masc expression in female embryos injected with two types of siRNAs that target Siwi (Siwi-1 and Siwi-2) or a control siRNA that targets GFP. Total RNA was isolated from female siRNA-injected embryos at 18 hpo and RT–qPCR was performed. The data shown are mean + s.d. The number at the base of each bar indicates the sample size of each group. Data were subjected to Kruskal–Wallis analysis with post hoc Dunn’s test. *P < 0.05. The expression levels of Siwi mRNA decreased to 23 and 44% after injecting Siwi-1 and Siwi-2 siRNAs, respectively, compared with that in GFP-siRNA-injected embryos. b, Knockdown of BmAgo3 mRNA in newly hatched larvae. The embryos were injected with BmAgo3 or GFP (control) siRNA. Total RNA was isolated from newly hatched larvae (at about 240 h post-injection) and RT–qPCR was performed. The data shown are mean + s.d. The number indicates the sample size of each group. *P < 0.05, one-sided Mann–Whitney test. c, Splicing of Bmdsx in newly hatched larvae that were injected with BmAgo3 siRNA. The Bmdsx splicing patterns were examined at about 240 h post-injection. The number indicates the sample size. The abbreviations are the same as in Fig. 2c. d, Masc expression in newly hatched larvae that were injected with BmAgo3 siRNA. Total RNA was isolated from siRNA-injected newly hatched larvae (at about 240 h post-injection) and RT–qPCR was performed. The data shown are mean + s.d. The number indicates the sample size of each group. *P < 0.05, one-sided Mann–Whitney test.
Extended Data Figure 8 Splicing of Bmdsx in Masc siRNA-injected embryos.
a, b, The Bmdsx splicing pattern was determined at 144 h (a) and 216 h (b) post-injection. The abbreviations are the same as in Fig. 2c. The number indicates the sample size.
Extended Data Figure 9 Functional analysis of the Fem piRNA-resistant Masc transcript.
a, Sequence of the Fem piRNA-resistant Masc (Masc-R) mRNA. Five nucleotide mutations that do not result in amino acid substitutions for the Masc protein are shown by red letters. The putative cleavage site by the Fem piRNA–Siwi complex is shown by the red line. b, RT–qPCR of Masc mRNA in cDNA-transfected BmN4 cells. BmN4 cells were transfected with Masc expression vectors or control vector. The Masc mRNA level was normalized to that of rp49. Data shown are means of duplicates. c, RT–qPCR of Masc piRNA in BmN4 cells transfected with Masc expression vectors or control vector. The Masc piRNA level was normalized to that of let-7. Data shown are means of duplicates. d, Identification of the cleavage site of exogenously introduced Masc. BmN4 cells were transfected with Masc expression vectors or control vector. Three days after transfection, zeocin (final concentration, 500 μg ml−1) was added to the medium. Six days after drug selection, the Masc mRNA-derived RNA fragment (shown by the red asterisk) expressed from the transfected plasmids was amplified by a modified RACE method. The fragment was cloned, sequenced, and identified as the Masc mRNA-derived one. The locations of the primers are shown by arrows. e, Effect of Masc transfection on the Bmdsx splicing in BmN4 cells. The splicing patterns of Bmdsx in stably transfected BmN4 cells (six days after drug selection) were examined by RT–PCR. The F and M indicate female- and male-type splicing of Bmdsx, respectively. Similar results were obtained in two independent experiments. f, Light microscopic observations of BmN4 cells stably transfected with Masc expression vectors or control vector (2 weeks after drug selection).
Extended Data Figure 10 Distribution of Masc-regulated genes throughout the silkworm genome.
a, b, The genome loci where Masc-regulated genes are located were identified using RNA-seq data from male (a) and female (b) embryos injected with control (siGFP) and Masc (siMasc) siRNAs (72 h post-injection).
Supplementary information
Supplementary Table 1
This file contains a list of oligonucleotides used in the study. (PDF 107 kb)
Supplementary Data
This file contains R computer code. (TXT 2 kb)
Source data
Rights and permissions
About this article
Cite this article
Kiuchi, T., Koga, H., Kawamoto, M. et al. A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature 509, 633–636 (2014). https://doi.org/10.1038/nature13315
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13315
This article is cited by
-
Accumulation of retrotransposons contributes to W chromosome differentiation in the willow beauty Peribatodes rhomboidaria (Lepidoptera: Geometridae)
Scientific Reports (2023)
-
Why put all your eggs in one basket? Evolutionary perspectives on the origins of monogenic reproduction
Heredity (2023)
-
First characterization of PIWI-interacting RNA clusters in a cichlid fish with a B chromosome
BMC Biology (2022)
-
Abundant small RNAs in the reproductive tissues and eggs of the honey bee, Apis mellifera
BMC Genomics (2022)
-
A male-specific doublesex isoform reveals an evolutionary pathway of sexual development via distinct alternative splicing mechanisms
Communications Biology (2022)