Abstract
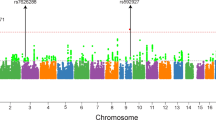
Schizophrenia, a devastating psychiatric disorder, has a prevalence of 0.5–1%, with high heritability (80–85%) and complex transmission1. Recent studies implicate rare, large, high-penetrance copy number variants in some cases2, but the genes or biological mechanisms that underlie susceptibility are not known. Here we show that schizophrenia is significantly associated with single nucleotide polymorphisms (SNPs) in the extended major histocompatibility complex region on chromosome 6. We carried out a genome-wide association study of common SNPs in the Molecular Genetics of Schizophrenia (MGS) case-control sample, and then a meta-analysis of data from the MGS, International Schizophrenia Consortium and SGENE data sets. No MGS finding achieved genome-wide statistical significance. In the meta-analysis of European-ancestry subjects (8,008 cases, 19,077 controls), significant association with schizophrenia was observed in a region of linkage disequilibrium on chromosome 6p22.1 (P = 9.54 × 10-9). This region includes a histone gene cluster and several immunity-related genes—possibly implicating aetiological mechanisms involving chromatin modification, transcriptional regulation, autoimmunity and/or infection. These results demonstrate that common schizophrenia susceptibility alleles can be detected. The characterization of these signals will suggest important directions for research on susceptibility mechanisms.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Accession codes
Primary accessions
DDBJ/GenBank/EMBL
Data deposits
Data have been deposited at dbGaP (http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap) under accessions phs000021.v2.p1 and phs000167.v1.p1, and the NIMH Center for Collaborative Genetic Studies on Mental Disorders (http://www.nimhgenetics.org) under studies 6, 29 and 29C.
Change history
06 August 2009
Author affiliation five was changed on 6 August 2009.
References
Tandon, R., Keshavan, M. S. & Nasrallah, H. A. Schizophrenia, “just the facts” what we know in 2008. 2. Epidemiology and etiology. Schizophr. Res. 102, 1–18 (2008)
Cook, E. H. & Scherer, S. W. Copy-number variations associated with neuropsychiatric conditions. Nature 455, 919–923 (2008)
Arnold, S. E., Talbot, K. & Hahn, C. G. Neurodevelopment, neuroplasticity, and new genes for schizophrenia. Prog. Brain Res. 147, 319–345 (2005)
The International Schizophrenia Consortium Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 10.1038/nature08185 (this issue)
Manolio, T. A., Brooks, L. D. & Collins, F. S. A. HapMap harvest of insights into the genetics of common disease. J. Clin. Invest. 118, 1590–1605 (2008)
Adegbola, A., Gao, H., Sommer, S. & Browning, M. A novel mutation in JARID1C/SMCX in a patient with autism spectrum disorder (ASD). Am. J. Med. Genet. A. 146A, 505–511 (2008)
Costa, E. et al. Reviewing the role of DNA (cytosine-5) methyltransferase overexpression in the cortical GABAergic dysfunction associated with psychosis vulnerability. Epigenetics 2, 29–36 (2007)
Kawasaki, H. & Iwamuro, S. Potential roles of histones in host defense as antimicrobial agents. Infect. Disord. Drug Targets 8, 195–205 (2008)
Malcherek, G. et al. The B7 homolog butyrophilin BTN2A1 is a novel ligand for DC-SIGN. J. Immunol. 179, 3804–3811 (2007)
Oksenberg, J. R., Baranzini, S. E., Sawcer, S. & Hauser, S. L. The genetics of multiple sclerosis: SNPs to pathways to pathogenesis. Nature Rev. Genet. 9, 516–526 (2008)
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007)
Huang, L. et al. Genotype-imputation accuracy across worldwide human populations. Am. J. Hum. Genet. 84, 235–250 (2009)
Stefansson, H. et al. Common variants conferring risk of schizophrenia. Nature 10.1038/nature08186 (this issue)
Wassink, T. H. et al. Evaluation of the chromosome 2q37.3 gene CENTG2 as an autism susceptibility gene. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 136B, 36–44 (2005)
Acknowledgements
We thank the study participants, and the research staff at the study sites. This study was supported by funding from the National Institute of Mental Health (USA) and the National Alliance for Research on Schizophrenia and Depression. Genotyping of part of the sample was supported by the Genetic Association Information Network (GAIN), and by The Paul Michael Donovan Charitable Foundation. Genotyping was carried out by the Center for Genotyping and Analysis at the Broad Institute of Harvard and MIT with support from the National Center for Research Resources (USA). The GAIN quality control team (G. R. Abecasis and J. Paschall) made important contributions to the project. We thank S. Purcell for assistance with PLINK.
Author Contributions J.S., D.F.L. and P.V.G. wrote the first draft of the paper. P.V.G., D.F.L., A.R.S., B.J.M., A.O., F.A., C.R.C., J.M.S., N.G.B., W.F.B., D.W.B., R.R.C. and R.F. oversaw the recruitment and clinical assessment of MGS participants, and the clinical aspects of the project and analysis. A.R.S., D.F.L. and P.V.G. performed database curation. D.F.L., J.S., I.P., F.D., P.A.H., A.S.W. and P.V.G. designed the analytical strategy and analysed the data. D.B.M. oversaw the Affymetrix 6.0 genotyping, and J.D., Y.Z., A.R.S. and P.V.G. performed the preparative genotyping and experimental work. J.R.O. contributed to the interpretation of data in the MHC/HLA region, and K.S.K. contributed to the approach to clinical data. P.V.G. coordinated the overall study. All authors contributed to the current version of the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
F.A. has received funds from Pfizer, Organon, and the Foundation for the National Institutes of Health. D.W.B. has received research support from Shire and Forest, has been on the speakers’ bureau for Pfizer, and has received consulting honoraria from Forest and Jazz.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary Tables S1-S19, Supplementary Figures S1-S11 and Supplementary References. (PDF 1142 kb)
Supplementary Data
This file contains data for the MGS GWAS and for the meta-analysis of MGS with ISC and SGENE data. (XLS 793 kb)
Supplementary Data
This file contains MGS GWAS results for 75 genes that have previously been considered candidate genes for schizophrenia, or closely related genes. (XLS 815 kb)
Supplementary Data
This file contains intensity plots for SNPs in the MHC region. (PDF 709 kb)
PowerPoint slides
Rights and permissions
About this article
Cite this article
Shi, J., Levinson, D., Duan, J. et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature 460, 753–757 (2009). https://doi.org/10.1038/nature08192
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08192
This article is cited by
-
Effects of MAP4K inhibition on neurite outgrowth
Molecular Brain (2023)
-
Significance of an altered lncRNA landscape in schizophrenia and cognition: clues from a case–control association study
European Archives of Psychiatry and Clinical Neuroscience (2023)
-
Schizophrenia in the genetic era: a review from development history, clinical features and genomic research approaches to insights of susceptibility genes
Metabolic Brain Disease (2023)
-
Chromatin modifier developmental pluripotency associated factor 4 (DPPA4) is a candidate gene for alcohol-induced developmental disorders
BMC Medicine (2022)
-
The schizophrenia-associated missense variant rs13107325 regulates dendritic spine density
Translational Psychiatry (2022)