Abstract
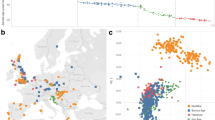
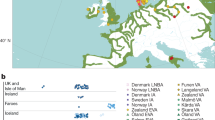
With the advent of dense maps of human genetic variation, it is now possible to detect positive natural selection across the human genome. Here we report an analysis of over 3 million polymorphisms from the International HapMap Project Phase 2 (HapMap2)1. We used ‘long-range haplotype’ methods, which were developed to identify alleles segregating in a population that have undergone recent selection2, and we also developed new methods that are based on cross-population comparisons to discover alleles that have swept to near-fixation within a population. The analysis reveals more than 300 strong candidate regions. Focusing on the strongest 22 regions, we develop a heuristic for scrutinizing these regions to identify candidate targets of selection. In a complementary analysis, we identify 26 non-synonymous, coding, single nucleotide polymorphisms showing regional evidence of positive selection. Examination of these candidates highlights three cases in which two genes in a common biological process have apparently undergone positive selection in the same population:LARGE and DMD, both related to infection by the Lassa virus3, in West Africa;SLC24A5 and SLC45A2, both involved in skin pigmentation4,5, in Europe; and EDAR and EDA2R, both involved in development of hair follicles6, in Asia.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature doi: 10.1038/nature06258 (this issue)
Sabeti, P. C. et al. Positive natural selection in the human lineage. Science 312, 1614–1620 (2006)
Kunz, S. et al. Posttranslational modification of α-dystroglycan, the cellular receptor for arenaviruses, by the glycosyltransferase LARGE is critical for virus binding. J. Virol. 79, 14282–14296 (2005)
Graf, J., Hodgson, R. & van Daal, A. Single nucleotide polymorphisms in the MATP gene are associated with normal human pigmentation variation. Hum. Mutat. 25, 278–284 (2005)
Lamason, R. L. et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 310, 1782–1786 (2005)
Botchkarev, V. A. & Fessing, M. Y. Edar signaling in the control of hair follicle development. J. Investig. Dermatol. Symp. Proc. 10, 247–251 (2005)
The International Haplotype Map Consortium. A haplotype map of the human genome. Nature 437, 1299–1320 (2005)
Sabeti, P. C. et al. Detecting recent positive selection in the human genome from haplotype structure. Nature 419, 832–837 (2002)
Voight, B. F., Kudaravalli, S., Wen, X. & Pritchard, J. K. A map of recent positive selection in the human genome. PLoS Biol. 4, e72 (2006)
Kimura, R., Fujimoto, A., Tokunaga, K. & Ohashi, J. A practical genome scan for population-specific strong selective sweeps that have reached fixation. PLoS ONE 2, e286 (2007)
Tang, K., Thornton, K. R. & Stoneking, M. A new approach for using genome scans to detect recent positive selection in the human genome. PLoS Biol. 5, e171 (2007)
Williamson, S. H. et al. Localizing recent adaptive evolution in the human genome. PLoS Genet. 3, e90 (2007)
Bersaglieri, T. et al. Genetic signatures of strong recent positive selection at the lactase gene. Am. J. Hum. Genet. 74, 1111–1120 (2004)
Teshima, K. M., Coop, G. & Przeworski, M. How reliable are empirical genomic scans for selective sweeps? 16, 702–712 Genome Res.. (2006)
Kuokkanen, M. et al. Transcriptional regulation of the lactase–phlorizin hydrolase gene by polymorphisms associated with adult-type hypolactasia. Gut 52, 647–652 (2003)
Miller, R. G. Simultaneous statistical inference XVI 299 (Springer, New York, 1981)
Soejima, M., Tachida, H., Ishida, T., Sano, A. & Koda, Y. Evidence for recent positive selection at the human AIM1 locus in a European population. Mol. Biol. Evol. 23, 179–188 (2006)
Richmond, J. K. & Baglole, D. J. Lassa fever: epidemiology, clinical features, and social consequences. Br. Med. J. 327, 1271–1275 (2003)
Colosimo, P. F. et al. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science 307, 1928–1933 (2005)
Rosenberg, N. A. et al. Genetic structure of human populations. Science 298, 2381–2385 (2002)
Chassaing, N., Bourthoumieu, S., Cossee, M., Calvas, P. & Vincent, M. C. Mutations in EDAR account for one-quarter of non-ED1-related hypohidrotic ectodermal dysplasia. Hum. Mutat. 27, 255–259 (2006)
Marti-Renom, M. A. et al. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29, 291–325 (2000)
Landau, M. et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33, W299–W302 (2005)
Xiao, T., Towb, P., Wasserman, S. A. & Sprang, S. R. Three-dimensional structure of a complex between the death domains of Pelle and Tube. Cell 99, 545–555 (1999)
Stephens, M., Smith, N. J. & Donnelly, P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68, 978–989 (2001)
Crawford, D. C. et al. Evidence for substantial fine-scale variation in recombination rates across the human genome. Nature Genet. 36, 700–706 (2004)
Schaffner, S. F. et al. Calibrating a coalescent simulation of human genome sequence variation. Genome Res. 15, 1576–1583 (2005)
Berglund, H. et al. The three-dimensional solution structure and dynamic properties of the human FADD death domain. J. Mol. Biol. 302, 171–188 (2000)
Huang, B., Eberstadt, M., Olejniczak, E. T., Meadows, R. P. & Fesik, S. W. NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain. Nature 384, 638–641 (1996)
Lasker, M. V., Gajjar, M. M. & Nair, S. K. Cutting edge: molecular structure of the IL-1R-associated kinase-4 death domain and its implications for TLR signaling. J. Immunol. 175, 4175–4179 (2005)
Liepinsh, E., Ilag, L. L., Otting, G. & Ibanez, C. F. NMR structure of the death domain of the p75 neurotrophin receptor. EMBO J. 16, 4999–5005 (1997)
Park, H. H. & Wu, H. Crystal structure of RAIDD death domain implicates potential mechanism of PIDDosome assembly. J. Mol. Biol. 357, 358–364 (2006)
Marti-Renom, M. A., Madhusudhan, M. S. & Sali, A. Alignment of protein sequences by their profiles. Protein Sci. 13, 1071–1087 (2004)
Kleywegt, G. J. Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr. D 52, 842–857 (1996)
DeLano, W. L. MacPyMOL: A PyMOL-based Molecular Graphics Application for MacOS X. (DeLano Scientific LLC, Palo Alto, California, USA, 2007)
Acknowledgements
P.C.S. is funded by a Burroughs Wellcome Career Award in the Biomedical Sciences and has been funded by the Damon Runyon Cancer Fellowship and the L’Oreal for Women in Science Award. We thank A. Schier, B. Voight, R. Roberts, M. Kreiger, A. Abzhanov, D. Degusta, M. Burnette, E. Lieberman, M. Daly, D. Altshuler, D. Reich, D. Lieberman and I. Woods for helpful discussions on our analysis and results. We also thank L. Ziaugra, D. Tabbaa and T. Rachupka for experimental assistance. This work was funded in part by grants from the National Human Genome Research Institute (to E.S.L.) and from the Broad Institute of MIT and Harvard.
Author Contributions P.C.S., P.V., B.F. and E.S.L. initiated the project. P.V., B.F. and P.C.S. developed key software. P.C.S., P.V., B.F., S.F.S., J.L., E.H., C.C., X.X., E.B., S.A.McC. and R.G. performed analysis. P.C.S., E.B. and E.H. performed experiments. P.C.S., E.S.L., P.V. and S.F.S. wrote the manuscript.
Author information
Authors and Affiliations
Consortia
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Lists of participants and affiliations appear at the end of the paper.
Supplementary information
Supplementary Information
The file contains Supplementary Methods, Supplementary Figures S1-S10 with Legends and Supplementary Tables 1-11. Supplementary Methods give details of LRH, iHS, XP-EHH, and localization analysis, including simulations and power calculations, Sweep software, ruling out confounders and details of identification of functional elements in the top candidates. Supplementary Tables illustrate power calculations for LRH, iHS, and XP-EHH, candidate regions and polymorphisms found from several analysis, fraction of SNPs predicted to be in the HapMap and dbSNP, as well as locations of copy number variants in the top candidates. Supplementary Figures show schematic of localization heuristic, power of LRH, iHS, and XP-EHH, top XP-EHH candidate,. localization of signal in LCT region, conservation, protein sequence and structure prediction, global variation, and examination of copy number variants for SLC24A5 and EDAR. (PDF 1482 kb)
Rights and permissions
About this article
Cite this article
Sabeti, P., Varilly, P., Fry, B. et al. Genome-wide detection and characterization of positive selection in human populations. Nature 449, 913–918 (2007). https://doi.org/10.1038/nature06250
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature06250
This article is cited by
-
Whole-genome resequencing of Chinese indigenous sheep provides insight into the genetic basis underlying climate adaptation
Genetics Selection Evolution (2024)
-
Deciphering climate resilience in Indian cattle breeds by selection signature analyses
Tropical Animal Health and Production (2024)
-
Genetic and cultural adaptations underlie the establishment of dairy pastoralism in the Tibetan Plateau
BMC Biology (2023)
-
Whole genome sequencing reveals signals of adaptive admixture in Creole cattle
Scientific Reports (2023)
-
SweepCluster: A SNP clustering tool for detecting gene-specific sweeps in prokaryotes
BMC Bioinformatics (2022)