Abstract
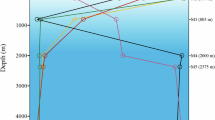
The most abundant class of bacterial ribosomal RNA genes detected in seawater DNA by gene cloning belongs to SAR11—an α-proteobacterial clade1. Other than indications of their prevalence in seawater, little is known about these organisms. Here we report quantitative measurements of the cellular abundance of the SAR11 clade in northwestern Sargasso Sea waters to 3,000 m and in Oregon coastal surface waters. On average, the SAR11 clade accounts for a third of the cells present in surface waters and nearly a fifth of the cells present in the mesopelagic zone. In some regions, members of the SAR11 clade represent as much as 50% of the total surface microbial community and 25% of the subeuphotic microbial community. By extrapolation, we estimate that globally there are 2.4 × 1028 SAR11 cells in the oceans, half of which are located in the euphotic zone. Although the biogeochemical role of the SAR11 clade remains uncertain, these data support the conclusion that this microbial group is among the most successful organisms on Earth.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Giovannoni, S. & Rappé, M. Microbial Ecology of the Oceans (ed. Kirchman, D. L.) 47–84 (John Wiley and Sons, New York, 2000)
Cho, B. C. & Azam, F. Major role of bacteria in biogeochemical fluxes in the ocean's interior. Nature 332, 441–443 (1988)
Fuhrman, J. A., Sleeter, T. D., Carlson, C. A. & Proctor, L. Dominance of bacterial biomass in the Sargasso Sea and its ecological implications. Mar. Ecol. Prog. Ser. 57, 207–217 (1989)
Buck, K. R., Chavez, F. P. & Campbell, L. Basin-wide distributions of living carbon components and the inverted trophic pyramid of the central gyre of the North Atlantic Ocean, summer 1993. Aquat. Microb. Ecol. 10, 238–298 (1996)
Carlson, C. A., Ducklow, H. W. & Sleeter, T. D. Stocks and dynamics of bacterioplankton in the northwestern Sargasso Sea. Deep-Sea Res. II 43, 491–516 (1996)
Karner, M. B., Delong, E. F. & Karl, D. M. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409, 507–510 (2001)
Fuhrman, J. A. & Ouverney, C. C. Marine microbial diversity studied via 16S rRNA sequences: cloning results from coastal waters and counting native archaea with fluorescent single cell probes. Aquat. Ecol. 32, 3–15 (1998)
Wintzingerode, F. V., Gobel, U. B. & Stackebrandt, E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21, 213–229 (1997)
Hicks, R. E., Amann, R. I. & Stahl, D. A. Dual staining of natural bacterioplankton with 4′6-diamidino-2-phenylindole and fluorescent oligonucleotide probes targeting kingdom-level 16S rRNA sequences. Appl. Environ. Microbiol. 58, 2158–2163 (1992)
DeLong, E. F., Wickham, G. S. & Pace, N. R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243, 1360–1363 (1989)
Amann, R. I., Krumholz, L. & Stahl, D. A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172, 762–770 (1990)
Glöckner, F. O. et al. An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst. Appl. Microbiol. 19, 403–406 (1996)
Frischer, M. E., Floriani, P. J. & Nierzwicki-Bauer, S. A. Differential sensitivity of 16S rRNA targeted oligonucleotide probes used for fluorescence in situ hybridization is a result of ribosomal higher order structure. Can. J. Microbiol. 42, 1061–1071 (1996)
Fuchs, B. M. et al. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 64, 4973–4982 (1998)
Fuchs, B. M., Glöckner, F. O., Wulf, J. & Amann, R. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 66, 3603–3607 (2000)
Lee, S., Malone, C. & Kemp, P. F. Use of multiple 16S rRNA-targeted fluorescent probes to increase signal strength and measure cellular RNA from natural planktonic bacteria. Mar. Ecol. Prog. Ser. 101, 193–201 (1993)
Schonhuber, W., Fuchs, B., Juretschko, S. & Amann, R. I. Improved sensitivity of whole-cell hybridization by the combination of horseradish peroxidase-labeled oligonucleotides and tyramide signal amplification. Appl. Environ. Microbiol. 63, 3268–3273 (1997)
Pernthaler, A., Preston, C. M., Pernthaler, J., DeLong, E. F. & Amann, R. Comparison of fluorescently labeled oligonucleotide and polynucleotide probes for the detection of pelagic marine bacteria and archaea. Appl. Environ. Microbiol. 68, 661–667 (2002)
Sieracki, M. E., Johnson, P. W. & Sieburth, J. M. Detection, enumeration, and sizing of planktonic bacteria by image-analyzed epifluorescence microscopy. Appl. Environ. Microbiol. 49, 799–810 (1985)
Porter, K. G. & Feig, Y. S. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25, 943–948 (1980)
Kemp, P. F., Lee, S. & LaRoche, J. Estimating the growth rate of slowly growing marine bacteria from RNA content. Appl. Environ. Microbiol. 59, 2594–2601 (1993)
Rappé, M. S., Connon, S. A., Vergin, K. L. & Giovannoni, S. J. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418, 630–633 (2002)
Cottrell, M. T. & Kirchman, D. L. Community composition of marine bacterioplankton determined by 16S rRNA gene clone libraries and fluorescence in situ hybridization. Appl. Environ. Microbiol. 66, 5116–5122 (2000)
Whitman, W. B., Coleman, D. C. & Wiebe, W. J. Prokaryotes: The unseen majority. Proc. Natl Acad. Sci. USA 95, 6578–6583 (1998)
Field, K. G. et al. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl. Environ. Microbiol. 63, 63–70 (1997)
Ludwig, W. et al. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19, 554–568 (1998)
Giovannoni, S. J., Britschgi, T. B., Moyer, C. L. & Field, K. G. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345, 60–63 (1990)
Acknowledgements
We thank R. Parsons, N. Nelson, the BATS scientific team and officers and crew of the RV Weatherbird II for help with collecting and processing samples. This work was supported by grants from Oregon State University, the Murdock Charitable Trust and the National Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Rights and permissions
About this article
Cite this article
Morris, R., Rappé, M., Connon, S. et al. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420, 806–810 (2002). https://doi.org/10.1038/nature01240
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature01240
This article is cited by
-
The microbial community of coral reefs: biofilm composition on artificial substrates under different environmental conditions
Marine Biology (2024)
-
Bacterial response to glucose addition: growth and community structure in seawater microcosms from North Pacific Ocean
Scientific Reports (2023)
-
Contrasting drivers of abundant phage and prokaryotic communities revealed in diverse coastal ecosystems
ISME Communications (2023)
-
Picoplankton diversity in an oligotrophic and high salinity environment in the central Adriatic Sea
Scientific Reports (2023)
-
Ubiquitous occurrence of a dimethylsulfoniopropionate ABC transporter in abundant marine bacteria
The ISME Journal (2023)