Abstract
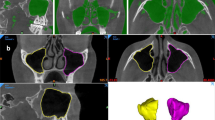
An accurate segmentation of the maxillary sinus (MS) is helpful for preoperative planning of dental implantation, diagnosis and evaluation of sinusitis, and validation of radiotherapy for sinus cancer. Many medical image segmentation models based on convolutional neural networks have achieved excellent performance, however, relied heavily on manual accurate labeling of training data. We propose an iterative learning method for MS segmentation with only bounding box supervision. First, a cone-beam computed tomography (CBCT) image is over-segmented into a set of superpixels and a feature extraction network is optimized to better extract multi-scale features of each small-size superpixel. Second, an improved graph convolutional network (IGCN) is developed to merge superpixel regions and improve the feature transformation ability of each node on a superpixel-wise graph. Finally, the iterative learning combined with the superpixel-conditional random field and IGCN makes pseudo labels gradually refine and close to fully supervised information. On a practical MS dataset, the proposed method achieves 90.5% in Dice similarity coefficient. Extending to the public dataset Promise12 for prostate MR image segmentation, it also performs well. The results show that our proposed method has good comprehensive weakly supervised segmentation performance and can narrow a gap between the bounding box and full supervision.











Similar content being viewed by others
Data availability
The data from stomatology hospital that support the findings of this study are available from the corresponding author (Fudong Zhu) upon reasonable request and with permission of stomatology hospital. The data from stomatology hospital are not publicly available, because they contain protected patient privacy information. Another data set is publicly available at the MICCAI2012 prostate MR segmentation challenge (https://promise12.grand-challenge.org/)
References
Achanta R, Shaji A, Smith K, Lucchi A, Fua P, Süsstrunk S (2012) SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Trans Pattern Anal Mach Intell 34(11):2274–2282. https://doi.org/10.1109/TPAMI.2012.120
Aksoy EA, Özden SU, Karaarslan E, Ünal ÖF, Tanyeri H (2014) Reliability of high-pitch ultra-low-dose paranasal sinus computed tomography for evaluating paranasal sinus anatomy and sinus disease. J Craniofac Surg 25(5):1801–1804. https://doi.org/10.1097/SCS.0000000000000966
Amanda FG, Thiago D, Cristina YM et al (2019) Development and validation of a formula based on maxillary sinus measurements as a tool for sex estimation: a cone beam computed tomography study. Int J Legal Med 133(4):1241–1249. https://doi.org/10.1007/s00414-018-1869-6
Amirkhani D, Bastanfard A (2019) Inpainted image quality evaluation based on saliency map features, in: 5th Iranian conference on signal processing and intelligent systems (ICSPIS), Shahrood, Iran, December 18-19, 2019: 1–6 https://doi.org/10.1109/ICSPIS48872.2019.9066140.
Andersen TN, Darvann TA, Murakami S et al (2018) Accuracy and precision of manual segmentation of the maxillary sinus in MR images - a method study. Br J Radiol 91(1085):1085. https://doi.org/10.1259/bjr.20170663
Bastanfard A, Amirkhani D, Mohammadi M (2022) Toward image super-resolution based on local regression and nonlocal means. Multimed Tools Appl 81:23473–23492. https://doi.org/10.1007/s11042-022-12584-x
Berberi A, Bouserhal L, Nader N, Assaf RB, Nassif NB, Bouserhal J, Salameh Z (2015) Evaluation of three-dimensional volumetric changes after sinus floor augmentation with mineralized cortical bone allograft. J Maxillofac Oral Surg 14(3):624–629. https://doi.org/10.1007/s12663-014-0736-3
Bui NL, Ong SH, Foong K (2015) Automatic segmentation of the nasal cavity and paranasal sinuses from cone-beam CT images. Int J Comput Assist Radiol Surg 10(8):1269–1277. https://doi.org/10.1007/s11548-014-1134-5
Cha JY, Yoon HI, Yeo IS et al (2021) Panoptic segmentation on panoramic radiographs: deep learning-based segmentation of various structures including maxillary sinus and mandibular canal. J Clin Med 10(12):2577. https://doi.org/10.3390/jcm10122577
Chen L C, Papandreou G, Kokkinos I, Murphy K, Yuille A L (2015) Semantic image segmentation with deep convolutional nets and fully connected CRFs. In: 3rd international conference on learning representations (ICLR), San Diego, CA, May 7-9, 2015
Choi H, Jeon KJ, Kim YH et al (2022) Deep learning-based fully automatic segmentation of the maxillary sinus on cone-beam computed tomographic images. Sci Rep 12:14009. https://doi.org/10.1038/s41598-022-18436-w
Cui F, Feng R, Wang L, Wei L (2021) Joint superpixel segmentation and graph convolutional network road extraction for high-resolution remote sensing imagery. In: IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Brussels, Belgium, July 11–16, 2021: 2178–2181. https://doi.org/10.1109/IGARSS47720.2021.9554635.
Demir UL, Akca ME, Ozpar R et al (2015) Anatomical correlation between existence of concha bullosa and maxillary sinus volume. Surg Radiol Anat 37(9):1093–1098. https://doi.org/10.1007/s00276-015-1459-y
Di S, Zhao Y, Liao M, Yang Z, Zeng Y (2022) Automatic liver tumor segmentation from CT images using hierarchical iterative superpixels and local statistical features. Expert Syst Appl 203:117347. https://doi.org/10.1016/j.eswa.2022.117347
Dosovitskiy A, Beyer L, Kolesnikov A et al (2021) An image is worth 16×16 words: transformers for image recognition at scale. In: 9th international conference on learning representations (ICRL), virtual event, Austria, May 3-7, 2021
Elineide S, Rodrigo MSV, Kelson RTA et al (2022) Semi-automatic segmentation of skin lesions based on superpixels and hybrid texture information. Med Image Anal 77:102363. https://doi.org/10.1016/j.media.2022.102363
Gugulothu V K, Balaji S (2023) An early prediction and classification of lung nodule diagnosis on CT images based on hybrid deep learning techniques. Multimed Tools Appl https://doi.org/10.1007/s11042-023-15802-2
Guilherme G, Menegatti P, Carrasco A et al (2018) Computed tomography-based volumetric tool for standardized measurement of the maxillary sinus. PLoS One 13(1):e0190770. https://doi.org/10.1371/journal.pone.0190770
Han K, Wang Y, Guo J, Tang Y, Wu E (2022) Vision GNN: an image is worth graph of nodes. In: Advances in Neural Information Processing Systems (NeurIPS), New Orleans, November 28–December 9, 2022, vol, 35, pp 8291–8303
He K, Zhang X, Ren S, Sun J (2016). Deep residual learning for image recognition. In: conference on computer vision and pattern recognition (CVPR), Las Vegas, NV, USA, June 27-30, 2016: 770-778. https://doi.org/10.1109/CVPR.2016.90
Hong Y, Zhang G, Wei B et al (2022) Weakly supervised semantic segmentation for skin cancer via CNN superpixel region response. Multimed Tools Appl 81:38409–38427. https://doi.org/10.1007/s11042-022-13606-4
Hu H, Ji D, Gan W et al (2020) Class-wise dynamic graph convolution for semantic segmentation. In: Computer Vision - ECCV 2020, 16th European Conference, Glasgow, UK, August 23–28, 2020, 12362: 1–17. https://doi.org/10.1007/978-3-030-58520-4_1
Huang Q, Huang Y, Luo Y, Yuan F, Li X (2020) Segmentation of breast ultrasound image with semantic classification of superpixels. Med Image Anal 61:101657. https://doi.org/10.1016/j.media.2020.101657
Hung KF, Ai QYH, King AD et al (2022) Automatic detection and segmentation of morphological changes of the maxillary sinus mucosa on cone-beam computed tomography images using a three-dimensional convolutional neural network 26: 3987–3998. https://doi.org/10.1007/s00784-021-04365-x
Imtiaz T, Rifat S, Fattah SA, Wahid KA (2020) Automated brain tumor segmentation based on multi-planar superpixel level features extracted from 3D MR images. IEEE Access 8:25335–25349. https://doi.org/10.1109/ACCESS.2019.2961630
Iqbal MJ, Bajwa UI, Gilanie G et al (2022) Automatic brain tumor segmentation from magnetic resonance images using superpixel-based approach. Multimed Tools Appl 81:38409–38427. https://doi.org/10.1007/s11042-022-13166-7
Jampani V, Sun D, Liu M-Y, Yang M-H, Kautz J (2018) Superpixel sampling networks. In: proceedings of the European conference on computer vision (ECCV), Munich, Germany, September 8-14, 2018: 363-380. https://doi.org/10.1007/978-3-030-01234-2\_22
Jinda-Apiraksa A, Ongt S H, Hiew L T et al (2010) A segmentation technique for maxillary sinus using the 3-D level set method. In: TENCON 2009–2009 IEEE region 10 conference, Singapore, January 23-26, 2009: 1–6. https://doi.org/10.1109/TENCON.2009.5396044.
Joachims T, Finley T, Yu C-NJ (2009) Cutting-plane training of structural SVMs. Mach Learn 77(1):27–59. https://doi.org/10.1007/s10994-009-5108-8
Jung SK, Lim HK, Lee S et al (2021) Deep active learning for automatic segmentation of maxillary sinus lesions using a convolutional neural network. Diagnostics 11(4):688. https://doi.org/10.3390/diagnostics11040688
Kervadec H, Dolz J, Wang S, Granger E, Ayed IB (2020) Bounding boxes for weakly supervised segmentation: global constraints get close to full supervision. In: Proceedings of the Third Conference on Medical Imaging with Deep Learning (PMLR), July 6–8, 2020, Montreal, QC, Canada, vol 121, pp 365–381
Keustermans W, Huysmans T, Schmelzer B et al (2019) Matlab toolbox for semi-automatic segmentation of the human nasal cavity based on active shape modeling. Comput Biol Med 105:27–38. https://doi.org/10.1016/j.compbiomed.2018.12.008
Khosravanian A, Rahmanimanesh M, Keshavarzi P, Mozaffari S (2021) Fast level set method for glioma brain tumor segmentation based on superpixel fuzzy clustering and lattice boltzmann method. Comput Methods Prog Biomed 198:105809. https://doi.org/10.1016/j.cmpb.2020.105809
Kipf TN, Welling M (2017) Semi-supervised classification with graph convolutional networks. In: international conference on learning representations (ICLR), Toulon, France, April 24-26, 2017
Lafferty J, McCallum A, Pereira F (2001) Conditional random fields: probabilistic models for segmenting and labeling sequence data. In: Proceedings of the 18th International Conference on Machine Learning (ICML), Williams College, Williamstown, MA, June 28–July 1, 2001, pp 282–289
Li Z, Chen J (2015) Superpixel segmentation using linear spectral clustering. In: proceedings of the IEEE conference on computer vision and pattern recognition (CVPR), Boston, MA, USA, June 7-12, 2015: 1356-1363. https://doi.org/10.1109/CVPR.2015.7298741
Li S, He J, Wang Y et al (2018) Blind CT image quality assessment via deep learning strategy: initial study. In: medical imaging 2018: image perception, observer performance, and technology assessment, Houston, Texas, United States, February 10-15, 2018, 10577: 105771A. https://doi.org/10.1117/12.2293240
Ling H, Gao J, Kar A, Chen W, Fidler S (2019) Fast interactive object annotation with curve-GCN. In: proceedings of the IEEE/CVF conference on computer vision and pattern recognition (CVPR), Long Beach, CA, USA, June 16-20, 2019: 5257-5266. https://doi.org/10.1109/CVPR.2019.00540
Ma F, Gao F, Sun J, Zhou H, Hussain A (2019) Attention graph convolution network for image segmentation in big SAR imagery data. Remote Sens 11(21):2586. https://doi.org/10.3390/rs11212586
Mahani GK, Li R, Evangelou N et al (2022) Bounding box based weakly supervised deep convolutional neural network for medical image segmentation using an uncertainty guided and spatially constrained loss. In: 19th international symposium on biomedical imaging (ISBI). Kolkata, India, march 28-31, 2022: 1-5. https://doi.org/10.1109/ISBI52829.2022.9761558
Martins A, Figueiredo M, Aguiar P, Smith N, Xing E (2015) AD3: alternating directions dual decomposition for map inference in graphical models. J Mach Learn Res 16:495–545. https://doi.org/10.5555/2789272.2789288
Milletari F, Navab N, Ahmadi SA (2016) V-net: fully convolutional neural networks for volumetric medical image segmentation. In: proceedings of the IEEE fourth international conference on 3D vision (3DV), Stanford, CA, USA, October 25-28, 2016: 565-571. https://doi.org/10.1109/3DV.2016.79
Morgan N, Van Gerven A, Smolders A et al (2022) Convolutional neural network for automatic maxillary sinus segmentation on cone-beam computed tomographic images. Sci Rep 12:7523. https://doi.org/10.1038/s41598-022-11483-3
Qu H, Wu P, Huang Q, Yi J, Yan Z et al (2020) Weakly supervised deep nuclei segmentation using partial points annotation in histopathology images. IEEE Trans Med Imaging 39(11):3655–3666. https://doi.org/10.1109/TMI.2020.3002244
Rajchl M, Lee MCH, Oktay O et al (2017) DeepCut: object segmentation from bounding box annotations using convolutional neural networks. IEEE Trans Med Imaging 36(2):674–683. https://doi.org/10.1109/TMI.2016.2621185
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation, in: medical image computing and computer-assisted intervention, MICCAI 2015, 18th international conference Munich, Germany, October 5-9, 2015: 234–241. https://doi.org/10.1007/978-3-319-24574-4\_28
Rother C, Kolmogorov V, Blake A (2004) “GrabCut”: interactive foreground extraction using iterated graph cuts. ACM Trans Graph 23(3):309–314. https://doi.org/10.1145/1015706.1015720
Shi J, Malik J (2000) Normalized cuts and image segmentation. IEEE Trans Pattern Anal Mach Intell 22(8):888–905. https://doi.org/10.1109/34.868688
Shi H, Scarfe WC, Farman AG (2006) Maxillary sinus 3D segmentation and reconstruction from cone beam CT data sets. Int J CARS 1(2):83–89. https://doi.org/10.1007/s11548-006-0041-9
Tang W, Qiu G (2021) Dense graph convolutional neural networks on 3D meshes for 3D object segmentation and classification. Image Vis Comput 114:10425. https://doi.org/10.1016/j.imavis.2021.104265
Tolstikhin I O, Houlsby N, Kolesnikov A et al (2021) MLP-Mixer: an all-MLP architecture for vision. In: Annual Conference on Neural Information Processing Systems (NeurIPS), December 6–14, 2021, virtual, pp 24261–24272
Valvano G, Leo A, Tsaftaris SA (2021) Learning to segment from scribbles using multi-scale adversarial attention gates. IEEE Trans Med Imaging 40(8):1990–2001. https://doi.org/10.1109/TMI.2021.3069634
Veličković P, Cucurull G, Casanova A, Romero A, Liò P, Bengio Y (2018) Graph attention networks. In international conference on learning representations (ICLR), Vancouver, BC, Canada, April 30 - May 3, 2018
Wei J, Hu Y, Li G, Cui S et al (2022) BoxPolyp: boost generalized polyp segmentation using extra coarse bounding box annotations. In: medical image computing and computer assisted intervention (MICCAI), Singapore, September 18-22, 2022, 13433: 67-77. https://doi.org/10.1007/978-3-031-16437-8\_7
Xu J, Wang S, Zhou Z, Liu J, Chen X (2020) Automatic CT image segmentation of maxillary sinus based on VGG network and improved V-net. Int J Comput Assist Radiol Surg 15(9):1457–1465. https://doi.org/10.1007/s11548-020-02228-6
Acknowledgments
This work was supported partly by National Natural Science Foundation of China (No. 62106225 and No. U20A20171), partly by Natural Science Foundation of Zhejiang Province (No. LY21F020027), partly by Zhejiang Province Public Welfare Technology Application Research Project (No. LGG20F020017), and partly by Key Programs for Science and Technology Development of Zhejiang Province (No. 2022C03113).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interests
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, X., Wang, K., Wang, C. et al. Iterative learning for maxillary sinus segmentation based on bounding box annotations. Multimed Tools Appl 83, 33263–33293 (2024). https://doi.org/10.1007/s11042-023-16544-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-16544-x