Abstract
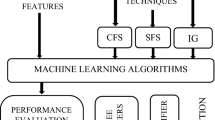
Accurate classifiers are vital to design precise computer aided diagnosis (CADx) systems. Classification performances of machine learning algorithms are sensitive to the characteristics of data. In this aspect, determining the relevant and discriminative features is a key step to improve performance of CADx. There are various feature extraction methods in the literature. However, there is no universal variable selection algorithm that performs well in every data analysis scheme. Random Forests (RF), an ensemble of trees, is used in classification studies successfully. The success of RF algorithm makes it eligible to be used as kernel of a wrapper feature subset evaluator. We used best first search RF wrapper algorithm to select optimal features of four medical datasets: colon cancer, leukemia cancer, breast cancer and lung cancer. We compared accuracies of 15 widely used classifiers trained with all features versus to extracted features of each dataset. The experimental results demonstrated the efficiency of proposed feature extraction strategy with the increase in most of the classification accuracies of the algorithms.









Similar content being viewed by others
References
Ming, L., and Zhi-Hua, Z., Improve computer-aided diagnosis with machine learning techniques using undiagnosed samples. Systems, man and cybernetics, part A: Systems and humans. IEEE Transactions on: 1088–1098, 2007.
Lee, M. C., Boroczky, L., Sungur-Stasik, K., Cann, A. D., Borczuk, A. C., Kawut, S. M., and Powell, C. A., A Two-step approach for feature selection and classifier ensemble construction in computer-aided diagnosis. In: Proceedings of the Proceedings of the 2008 21st IEEE International Symposium on Computer-Based Medical Systems, 2008.
Sun, S., Zhang, C., and Zhang, D., An experimental evaluation of ensemble methods for EEG signal classification. Pattern Recogn. Lett.: 2157–2163, 2007.
Ko, A. H. R., Sabourin, R., and de Souza Britt, A., Combining diversity and classification accuracy for ensemble selection in random subspaces. City, 2006.
Schapire, R., The boosting approach to machine learning: An overview. Nonlinear estimation and classification: Springer, 2003.
Breiman, L., Bagging predictors. Mach. Learn.: 123–140, 1996.
Polikar, R., Ensemble based systems in decision making. IEEE Circuits Syst. Mag.: 21–45, 2006.
Katz, J. D., Mamyrova, G., Guzhva, O., and Furmark, L., Random forests classification analysis for the assessment of diagnostic skill. Am. J. Med. Qual.: 149–153, 2010.
Huazhen, W., Chengde, L., Yanqing, P., and Xueqin, H., Application of improved random forest variables importance measure to traditional Chinese chronic gastritis diagnosis. City, 2008.
Ramírez, J., Górriz, J. M., Segovia, F., Chaves, R., Salas-Gonzalez, D., López, M., Álvarez, I., and Padilla, P., Computer aided diagnosis system for the Alzheimer’s disease based on partial least squares and random forest SPECT image classification. Neurosci. Lett.: 99–103, 2010.
Ozcift, A., Random forests ensemble classifier trained with data resampling strategy to improve cardiac arrhythmia diagnosis. Comput. Biol. Med., 2011. doi:10.1016/j.compbiomed.2011.03.001.
Yang, F., Wang, H., Mi, H., Lin, C., and Cai, W., Using random forest for reliable classification and cost-sensitive learning for medical diagnosis. BMC Bioinform. 10(Suppl 1):S22, 2010.
Nguyen, H.-N., Vu, T.-N., Ohn, S.-Y., Park, Y.-M., Han, M., and Kim, C., Feature elimination approach based on random forest for cancer diagnosis: Springer, City, 2006.
Janecek, A., and Wilfried, G., On the relationship between feature selection and classification accuracy. JMLR: Workshop Conf Proc: 90–105, 2008.
Martinez, A. M., and Manli, Z., Where are linear feature extraction methods applicable? Pattern analysis and machine intelligence. IEEE Transactions on: 1934–1944, 2005.
Saeys, Y., Inza, I., and Larrañaga, P., A review of feature selection techniques in bioinformatics. Bioinformatics: 2507–2517, 2007.
Kohavi, R., and John, G. H., Wrappers for feature subset selection. Artif. Intell.: 273–324, 1997.
Guyon, I. (Ed.), Feature extraction, foundations and applications. Stud. Fuzziness Soft Comput: 119–135, 2006.
Thongkam, J., Guandong, X., and Yanchun, Z., AdaBoost algorithm with random forests for predicting breast cancer survivability. City, 2008.
Chan, J. C.-W., and Paelinckx, D., Evaluation of random forest and adaboost tree-based ensemble classification and spectral band selection for ecotope mapping using airborne hyperspectral imagery. Remote Sens. Environ.: 2999–3011, 2008.
Alon, U. et al., Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc. Natl. Acad. Sci. U. S. A.: 6745–6750, 1999.
Golub, T. R., Slonim, D. K., and Tamayo, P., Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc. Natl Acad. Sci. 96:6745–6750, 1999.
Estrela da Silva, J., Marques de Sá, J., and Jossinet, J., Classification of breast tissue by electrical impedance spectroscopy. Med. Biol. Eng. Comput.: 26–30, 2000.
Hong, Z. Q., and Yang, J. Y., Optimal discriminant plane for a small number of samples and design method of classifier on the plane. Pattern Recognit. 24(4):317–324, 1991.
Hall, M. et al., The WEKA data mining software: An update. SIGKDD Explor. Newsl. 11:10–18, 2009.
Viswanathan, M., Measurement error and research design: Sage Publications: 44–60, 2005.
David, A., Comparison of classification accuracy using Cohen’s weighted Kappa. Expert Syst. Appl.: 825–832, 2008.
Kohavi, R., A study of cross-validation and bootstrap for accuracy estimation and model selection, In: Proceedings of the 14th international joint conference on Artificial intelligence: Morgan Kaufmann Publishers Inc.: 1137–1143, 1995.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ozcift, A. Enhanced Cancer Recognition System Based on Random Forests Feature Elimination Algorithm. J Med Syst 36, 2577–2585 (2012). https://doi.org/10.1007/s10916-011-9730-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10916-011-9730-1