Abstract
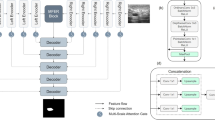
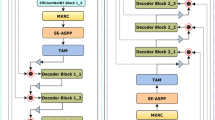
Medical image segmentation plays a vital role in assisting doctors to quickly and accurately identify pathological areas in medical images, which is essential for effective diagnosis and treatment planning. Although current architectures built by integrating CNNs and Transformers have achieved impressive results, they are still constrained by the limitations of fusion methods and the computational complexity of Transformers. To address these restrictions, we introduce a dual triple attention module designed to encourage selective modeling of image features, thereby enhancing the overall performance of the segmentation process. We design the feature extraction module based on CNN and VMamba for both local and global feature extraction. The CNN part includes regular convolutions and dilated convolutions, and we use the Visual State Space Block from VMamba instead of Transformers to reduce computational complexity. The entire network is connected through new skip connections. Experimental results show that the proposed model achieved DSC scores of 92.34% and 82.16% on the ACDC and Synapse datasets, respectively. Meanwhile, the FLOPs and parameters are also superior to other state-of-the-art methods.








Similar content being viewed by others
References
Ma, J., Zhang, Y., Gu, S., Zhu, C., Ge, C., Zhang, Y., An, X., Wang, C., Wang, Q., Liu, X., Cao, S., Zhang, Q., Liu, S., Wang, Y., Li, Y., He, J., Yang, X.: Abdomenct-1k: Is abdominal organ segmentation a solved problem? IEEE Trans. Pattern Anal. Mach. Intell. 44(10), 6695–6714 (2022). https://doi.org/10.1109/TPAMI.2021.3100536
Ronneberger, O., Fischer, P., Brox, T.: U-net: convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, pp. 234–241. Springer (2015)
Isensee, F., Jaeger, P.F., Kohl, S.A., Petersen, J., Maier-Hein, K.H.: nnu-net: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18(2), 203–211 (2021)
Agarwal, R., Ghosal, P., Murmu, N., Nandi, D.: Spiking neural network in computer vision: techniques, tools and trends. In: Borah, S., Gandhi, T.K., Piuri, V. (eds.) Advanced Computational and Communication Paradigms, pp. 201–209. Springer, Singapore (2023)
Li, B., Wang, Y., Xu, Y., Wu, C.: Dsst: a dual student model guided student-teacher framework for semi-supervised medical image segmentation. Biomed. Signal Process. Control 90, 105890 (2024). https://doi.org/10.1016/j.bspc.2023.105890
Wu, H., Pan, J., Li, Z., Wen, Z., Qin, J.: Automated skin lesion segmentation via an adaptive dual attention module. IEEE Trans. Med. Imaging 40(1), 357–370 (2021). https://doi.org/10.1109/TMI.2020.3027341
Peng, Z., Huang, W., Gu, S., Xie, L., Wang, Y., Jiao, J., Ye, Q.: Conformer: local features coupling global representations for visual recognition. In: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 367–376 (2021)
Guo, J., Han, K., Wu, H., Tang, Y., Chen, X., Wang, Y., Xu, C.: Cmt: convolutional neural networks meet vision transformers. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 12175–12185 (2022)
Ma, J., Li, F., Wang, B.: U-mamba: Enhancing long-range dependency for biomedical image segmentation. CoRR. arxiv:2401.04722 (2024)
Azad, R., Arimond, R., Aghdam, E.K., Kazerouni, A., Merhof, D.: DAE-former: dual attention-guided efficient transformer for medical image segmentation. In: Rekik, I., Adeli, E., Park, S.H., Cintas, C., Zamzmi, G. (eds.) PRIME, Canada. Lecture Notes in Computer Science, vol. 14277, pp. 83–95. Springer (2023). https://doi.org/10.1007/978-3-031-46005-0_8
Ghosal, P., Reddy, S., Sai, C., Pandey, V., Chakraborty, J., Nandi, D.: A deep adaptive convolutional network for brain tumor segmentation from multimodal MR images. In: TENCON 2019 - 2019 IEEE Region 10 Conference (TENCON), pp. 1065–1070 (2019). https://doi.org/10.1109/TENCON.2019.8929402
Chen, J., Lu, Y., Yu, Q., Luo, X., Adeli, E., Wang, Y., Lu, L., Yuille, A.L., Zhou, Y.: Transunet: Transformers make strong encoders for medical image segmentation. CoRR. arxiv:2102.04306 (2021)
Huang, X., Deng, Z., Li, D., Yuan, X., Fu, Y.: Missformer: an effective transformer for 2d medical image segmentation. IEEE Trans. Med. Imaging 42(5), 1484–1494 (2023). https://doi.org/10.1109/TMI.2022.3230943
Zhou, H., Guo, J., Zhang, Y., Han, X., Yu, L., Wang, L., Yu, Y.: nnformer: Volumetric medical image segmentation via a 3d transformer. IEEE Trans. Image Process. 32, 4036–4045 (2023). https://doi.org/10.1109/TIP.2023.3293771
Lin, G., Chen, L.: A multi-scale fusion network with transformer for medical image segmentation. In: 2023 3rd International Conference on Neural Networks, Information and Communication Engineering (NNICE), pp. 224–228 (2023). IEEE
Xu, L., Chen, M., Cheng, Y., Shao, P., Shen, S., Yao, P., Xu, R.X.: MCPA: multi-scale cross perceptron attention network for 2d medical image segmentation. CoRR. arxiv: 2307.14588 (2023)
Ke, Y., Yu, S., Wang, Z., Li, Y.: ECSFF: Exploring efficient cross-scale feature fusion for medical image segmentation. In: 2023 28th International Conference on Automation and Computing (ICAC), pp. 1–6 (2023). IEEE
Wang, H., Cao, P., Wang, J., Zaiane, O.R.: Uctransnet: rethinking the skip connections in U-net from a channel-wise perspective with transformer. In: Proceedings of the AAAI Conference on Artificial Intelligence, vol. 36, pp. 2441–2449 (2022)
Gu, A., Dao, T.: Mamba: Linear-time sequence modeling with selective state spaces. CoRR arxiv:2312.00752 (2023) https://doi.org/10.48550/ARXIV.2312.00752
Zhu, L., Liao, B., Zhang, Q., Wang, X., Liu, W., Wang, X.: Vision mamba: Efficient visual representation learning with bidirectional state space model. arXiv preprint arXiv:2401.09417 (2024)
Liu, Y., Tian, Y., Zhao, Y., Yu, H., Xie, L., Wang, Y., Ye, Q., Liu, Y.: Vmamba: Visual state space model. CoRR. arxiv:2401.10166 (2024) https://doi.org/10.48550/ARXIV.2401.10166
Ruan, J., Xiang, S.: Vm-unet: vision mamba unet for medical image segmentation. CoRR. arxiv:2402.02491 (2024)
Liu, J., Yang, H., Zhou, H., Xi, Y., Yu, L., Yu, Y., Liang, Y., Shi, G., Zhang, S., Zheng, H., Wang, S.: Swin-umamba: mamba-based unet with imagenet-based pretraining. CoRR. arxiv:2402.03302 (2024)
Wang, L., Li, D., Dong, S., Meng, X., Zhang, X., Hong, D.: Pyramidmamba: Rethinking pyramid feature fusion with selective space state model for semantic segmentation of remote sensing imagery. arXiv preprint arXiv:2406.10828 (2024)
Chen, K., Chen, B., Liu, C., Li, W., Zou, Z., Shi, Z.: RSMamba: remote sensing image classification with state space model. IEEE Geosci. Remote Sens. Lett. (2024)
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., Wojna, Z.: Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2818–2826 (2016)
Azad, R., Heidari, M., Yilmaz, K., Hüttemann, M., Karimijafarbigloo, S., Wu, Y., Schmeink, A., Merhof, D.: Loss functions in the era of semantic segmentation: A survey and outlook. CoRR. arxiv:2312.05391 (2023) https://doi.org/10.48550/ARXIV.2312.05391
Bernard, O., Lalande, A., Zotti, C., Cervenansky, F., Yang, X., Heng, P.-A., Cetin, I., Lekadir, K., Camara, O., Ballester, M.A.G., et al.: Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: is the problem solved? IEEE Trans. Med. Imaging 37(11), 2514–2525 (2018)
Fu, S., Lu, Y., Wang, Y., Zhou, Y., Shen, W., Fishman, E., Yuille, A.: Domain adaptive relational reasoning for 3d multi-organ segmentation. In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, pp. 656–666. Springer (2020)
Schlemper, J., Oktay, O., Schaap, M., Heinrich, M., Kainz, B., Glocker, B., Rueckert, D.: Attention gated networks: learning to leverage salient regions in medical images. Med. Image Anal. 53, 197–207 (2019)
Xu, G., Zhang, X., He, X., Wu, X.: LeViT-UNet: make faster encoders with Transformer for medical image segmentation. In: Liu, Q., Wang, H., Ma, Z., Zheng, W., Zha, H., Chen, X., Wang, L., Ji, R. (eds.) Pattern Recognition and Computer Vision, pp. 42–53. Springer, Singapore (2024)
Cao, H., Wang, Y., Chen, J., Jiang, D., Zhang, X., Tian, Q., Wang, M.: Swin-unet: Unet-like pure transformer for medical image segmentation. In: European Conference on Computer Vision, pp. 205–218. Springer (2022)
Azad, R., Al-Antary, M.T., Heidari, M., Merhof, D.: Transnorm: transformer provides a strong spatial normalization mechanism for a deep segmentation model. IEEE Access 10, 108205–108215 (2022)
Azad, R., Heidari, M., Shariatnia, M., Aghdam, E.K., Karimijafarbigloo, S., Adeli, E., Merhof, D.: Transdeeplab: convolution-free transformer-based Deeplab V3+ for medical image segmentation. In: International Workshop on PRedictive Intelligence In MEdicine, pp. 91–102 (2022)
Heidari, M., Kazerouni, A., Soltany, M., Azad, R., Aghdam, E.K., Cohen-Adad, J., Merhof, D.: Hiformer: hierarchical multi-scale representations using transformers for medical image segmentation. In: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, pp. 6202–6212 (2023)
Rahman, M.M., Marculescu, R.: Medical image segmentation via cascaded attention decoding. In: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, pp. 6222–6231 (2023)
Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn, D., Zhai, X., Unterthiner, T., Dehghani, M., Minderer, M., Heigold, G., Gelly, S., Uszkoreit, J., Houlsby, N.: An image is worth 16x16 words: transformers for image recognition at scale. In: 9th International Conference on Learning Representations (2021)
Hatamizadeh, A., Tang, Y., Nath, V., Yang, D., Myronenko, A., Landman, B., Roth, H.R., Xu, D.: Unetr: transformers for 3d medical image segmentation. In: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, pp. 574–584 (2022)
Shaker, A.M., Maaz, M., Rasheed, H., Khan, S., Yang, M.-H., Khan, F.S.: Unetr++: delving into efficient and accurate 3d medical image segmentation. IEEE Trans. Medical Imaging (2024). https://doi.org/10.1109/TMI.2024.3398728
Azad, R., Jia, Y., Aghdam, E.K., Cohen-Adad, J., Merhof, D.: Enhancing medical image segmentation with transception: a multi-scale feature fusion approach. CoRR. arxiv:2301.10847 (2023) https://doi.org/10.48550/ARXIV.2301.10847
Zhu, L., Wang, X., Ke, Z., Zhang, W., Lau, R.W.: Biformer: vision transformer with bi-level routing attention. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 10323–10333 (2023)
Dai, J., Liu, T., Torigian, D.A., Tong, Y., Han, S., Nie, P., Zhang, J., Li, R., Xie, F., Udupa, J.K.: Ga-net: a geographical attention neural network for the segmentation of body torso tissue composition. Med. Image Anal. 91, 102987 (2024). https://doi.org/10.1016/j.media.2023.102987
Funding
This work was supported by the Zhejiang Provincial Natural Science Foundation of China under Grant LQ23F020021.
Author information
Authors and Affiliations
Contributions
Conceptualization: Qiaohong Chen; Methodology: Qiaohong Chen; Formal analysis and investigation: Jing Li, Xian Fang; Writing - original draft preparation: Jing Li; Writing - review and editing: Qiaohong Chen, Xian Fang; Funding acquisition: Xian Fang; Resources: Xian Fang; Supervision: Xian Fang.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no competing of interest.
Ethical approval
Not applicable.
Additional information
Communicated by Haojie Li.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, Q., Li, J. & Fang, X. Dual triple attention guided CNN-VMamba for medical image segmentation. Multimedia Systems 30, 275 (2024). https://doi.org/10.1007/s00530-024-01498-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00530-024-01498-3