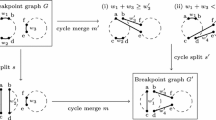
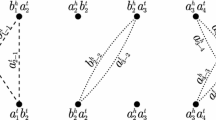
Abstract
The gene order on a chromosome is a necessary data for most comparative genomics studies, but in many cases only partial orders can be obtained by current genetic mapping techniques. The Minimum Breakpoint Linearization Problem aims at constructing a total order from this partial knowledge, such that the breakpoint distance to a reference genome is minimized. In this paper, we first expose a flaw in two algorithms formerly known for this problem [4,2]. We then present a new modeling for this problem, and use it to design three approximation algorithms, with ratios resp. O(log(k)loglog(k)), O(log2(|X|)) and m 2 + 4m − 4, where k is the optimal breakpoint distance we look for, |X| is upper bounded by the number of pair of genes for which the partial order is in contradiction with the reference genome, and m is the number of genetic maps used to create the input partial order.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Blin, G., Blais, E., Hermelin, D., Guillon, P., Blanchette, M., El-Mabrouk, N.: Gene maps linearization using genomic rearrangement distances. Journal of Computational Biology 14(4), 394–407 (2007)
Chen, X., Cui, Y.: An approximation algorithm for the minimum breakpoint linearization problem. IEEE/ACM Trans. Comput. Biology Bioinform. 6(3), 401–409 (2009)
Even, G., Naor, J., Schieber, B., Sudan, M.: Approximating minimum feedback sets and multi-cuts in directed graphs. In: Balas, E., Clausen, J. (eds.) IPCO 1995. LNCS, vol. 920, pp. 14–28. Springer, Heidelberg (1995)
Fu, Z., Jiang, T.: Computing the breakpoint distance between partially ordered genomes. J. Bioinformatics and Computational Biology 5(5), 1087–1101 (2007)
Yap, I.V., Schneider, D., Kleinberg, J., Matthews, D., Cartinhourb, S., McCouch, S.R.: A graph-theoretic approach to comparing and integrating genetic, physical and sequence-based maps. Genetics 165(4), 2235–2247 (2003)
Zheng, C., Lenert, A., Sankoff, D.: Reversal distance for partially ordered genomes. In: ISMB (Supplement of Bioinformatics), pp. 502–508 (2005)
Zheng, C., Sankoff, D.: Genome rearrangements with partially ordered chromosomes. In: Wang, L. (ed.) COCOON 2005. LNCS, vol. 3595, pp. 52–62. Springer, Heidelberg (2005)
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2010 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Bulteau, L., Fertin, G., Rusu, I. (2010). Revisiting the Minimum Breakpoint Linearization Problem. In: Kratochvíl, J., Li, A., Fiala, J., Kolman, P. (eds) Theory and Applications of Models of Computation. TAMC 2010. Lecture Notes in Computer Science, vol 6108. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-13562-0_16
Download citation
DOI: https://doi.org/10.1007/978-3-642-13562-0_16
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-13561-3
Online ISBN: 978-3-642-13562-0
eBook Packages: Computer ScienceComputer Science (R0)