Abstract
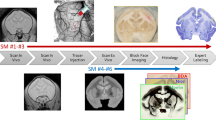
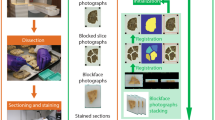
Immunohistochemistry is widely used as a gold standard to inspect tissues, characterize their structure and detect pathological alterations. As such, the joint analysis of histological images and other imaging modalities (MRI, PET) is of major interest to interpret these physical signals and establish their correspondence with the biological constitution of the tissues. However, it is challenging to provide a meaningful characterization of the signal specificity. In this paper, we propose an integrated method to quantitatively evaluate the discriminative power of imaging modalities. This method was validated using a macaque brain dataset containing: 3 immunohistochemically stained and 1 histochemically stained series, 1 photographic volume and 1 in vivo T2 weighted MRI. First, biological regions of interest (ROIs) were automatically delineated from histological sections stained for markers of interest and mapped on the target non-specific modalities through co-registration. These non-overlapping ROIs were considered ground truth for later classification. Voxels were evenly split in training and testing sets for a logistic regression model. The statistical significance of resulting accuracy scores was evaluated through null distribution simulations. Such an approach could be of major interest to assess relevant biological characteristics from various imaging modalities.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Mega, M.S., Chen, S.S., Thompson, P.M., Woods, R.P., Karaca, T.J., Tiwari, A., Vinters, H.V., Small, G.W., Toga, A.W.: Mapping histology to metabolism: coregistration of stained whole-brain sections to premortem PET in Alzheimer’s disease. Neuroimage 5, 147–153 (1997)
Piert, M., Park, H., Khan, A., Siddiqui, J., Hussain, H., Chenevert, T., Wood, D., Johnson, T., Shah, R.B., Meyer, C.: Detection of aggressive primary prostate cancer with 11C-choline PET/CT using multimodality fusion techniques. J. Nucl. Med. 50, 1585–1593 (2009)
Lavisse, S., Guillermier, M., Hérard, A.-S., Petit, F., Delahaye, M., Van Camp, N., Ben Haim, L., Lebon, V., Remy, P., Dollé, F., Delzescaux, T., Bonvento, G., Hantraye, P., Escartin, C.: Reactive astrocytes overexpress TSPO and are detected by TSPO positron emission tomography imaging. J. Neurosci. 32, 10809–10818 (2012)
Bürgel, U., Schormann, T., Schleicher, A., Zilles, K.: Mapping of histologically identified long fiber tracts in human cerebral hemispheres to the MRI volume of a reference brain: position and spatial variability of the optic radiation. Neuroimage 10, 489–499 (1999)
Ourselin, S., Roche, A., Subsol, G., Pennec, X., Ayache, N.: Reconstructing a 3D structure from serial histological sections. Image Vis. Comput. 19, 25–31 (2001)
Bardinet, E., Ourselin, S., Dormont, D., Malandain, G., Tandé, D., Parain, K., Ayache, N., Yelnik, J.: Co-registration of histological, optical and MR data of the human brain. In: Dohi, T., Kikinis, R. (eds.) MICCAI 2002, Part I. LNCS, vol. 2488, pp. 548–555. Springer, Heidelberg (2002)
Malandain, G., Bardinet, E., Nelissen, K., Vanduffel, W.: Fusion of autoradiographs with an MR volume using 2-D and 3-D linear transformations. Neuroimage 23, 111–127 (2004)
Lebenberg, J., Hérard, A., Dubois, A., Dhenain, M., Hantraye, P., Delzescaux, T.: A combination of atlas-based and voxel-wise approaches to analyze metabolic changes in autoradiographic data from Alzheimer’s mice. Neuroimage 57, 1447–1457 (2011)
Oh, S.W., Harris, J.A., Ng, L., Winslow, B., Cain, N., Mihalas, S., Wang, Q., Lau, C., Kuan, L., Henry, A.M., Mortrud, M.T., Ouellette, B., Nguyen, T.N., Sorensen, S.A., Slaughterbeck, C.R., Wakeman, W., Li, Y., Feng, D., Ho, A., Nicholas, E., Hirokawa, K.E., Bohn, P., Joines, K.M., Peng, H., Hawrylycz, M.J., Phillips, J.W., Hohmann, J.G., Wohnoutka, P., Gerfen, C.R., Koch, C., Bernard, A., Dang, C., Jones, A.R., Zeng, H.: A mesoscale connectome of the mouse brain. Nature 508, 207–214 (2014)
Amunts, K., Lepage, C., Borgeat, L., Mohlberg, H., Dickscheid, T., Rousseau, M.-É., Bludau, S., Bazin, P.-L., Lewis, L.B., Oros-Peusquens, A.-M., Shah, N.J., Lippert, T., Zilles, K., Evans, A.C.: BigBrain: an ultrahigh-resolution 3D human brain model. Science 340, 1472–1475 (2013)
Mayerich, D., Abbott, L., McCormick, B.: Knife-edge scanning microscopy for imaging and reconstruction of three-dimensional anatomical structures of the mouse brain. J. Microsc. 231, 134–143 (2008)
Ragan, T., Kadiri, L.R., Venkataraju, K.U., Bahlmann, K., Sutin, J., Taranda, J., Arganda-Carreras, I., Kim, Y., Seung, H.S., Osten, P.: Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat. Methods 9, 255–258 (2012)
Ertürk, A., Becker, K., Jährling, N., Mauch, C.P., Hojer, C.D., Egen, J.G., Hellal, F., Bradke, F., Sheng, M., Dodt, H.-U.: Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat. Protoc. 7, 1983–1995 (2012)
Chung, K., Wallace, J., Kim, S.-Y., Kalyanasundaram, S., Andalman, A.S., Davidson, T.J., Mirzabekov, J.J., Zalocusky, K.A., Mattis, J., Denisin, A.K., Pak, S., Bernstein, H., Ramakrishnan, C., Grosenick, L., Gradinaru, V., Deisseroth, K.: Structural and molecular interrogation of intact biological systems. Nature 497, 332–337 (2013)
Renier, N., Wu, Z., Simon, D.J., Yang, J., Ariel, P., Tessier-Lavigne, M.: iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910 (2014)
Susaki, E.A., Tainaka, K., Perrin, D., Kishino, F., Tawara, T., Watanabe, T.M., Yokoyama, C., Onoe, H., Eguchi, M., Yamaguchi, S., Abe, T., Kiyonari, H., Shimizu, Y., Miyawaki, A., Yokota, H., Ueda, H.R.: Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell 157, 726–739 (2014)
Osechinskiy, S., Kruggel, F.: Quantitative comparison of high-resolution MRI and myelin-stained histology of the human cerebral cortex. In: 31st Annual International Conference of the IEEE Engineering in Medicine and Biology Society, pp. 85–89 (2009)
Bol, K., Haeck, J.C., Alic, L., Niessen, W.J., de Jong, M., Bernsen, M., Veenland, J.F.: Quantification of DCE-MRI: a validation of three techniques with 3D-histology. In: 2012 9th IEEE International Symposium on Biomedical Imaging (ISBI), pp. 1044–1047. IEEE (2012)
van Engelen, A., Niessen, W.J., Klein, S., Groen, H.C., Verhagen, H.J.M., Wentzel, J.J., van der Lugt, A., de Bruijne, M.: Supervised in-vivo plaque characterization incorporating class label uncertainty. In: 2012 9th IEEE International Symposium on Biomedical Imaging (ISBI), pp. 246–249. IEEE (2012)
Stille, M., Smith, E.J., Crum, W.R., Modo, M.: 3D reconstruction of 2D fluorescence histology images and registration with in vivo MR images: application in a rodent stroke model. J. Neurosci. Methods 219, 27–40 (2013)
Coquery, N., Francois, O., Lemasson, B., Debacker, C., Farion, R., Rémy, C., Barbier, E.L.: Microvascular MRI and unsupervised clustering yields histology-resembling images in two rat models of glioma. J. Cereb. Blood Flow Metab. 34, 1354–1362 (2014)
Goubran, M., Hammond, R.R., de Ribaupierre, S., Burneo, J.G., Mirsattari, S., Steven, D.A., Parrent, A.G., Peters, T.M., Khan, A.R.: Magnetic resonance imaging and histology correlation in the neocortex in temporal lobe epilepsy. Ann. Neurol. 77, 237–250 (2015)
Lindvall, O., Kokaia, Z.: Stem cells for the treatment of neurological disorders. Nature 441, 1094–1096 (2006)
Ross, C.A., Akimov, S.S.: Human-induced pluripotent stem cells: potential for neurodegenerative diseases. Hum. Mol. Genet. 23, R17–R26 (2014)
Palfi, S., Condé, F., Riche, D., Brouillet, E., Dautry, C., Mittoux, V., Chibois, A., Peschanski, M., Hantraye, P.: Fetal striatal allografts reverse cognitive deficits in a primate model of Huntington disease. Nat. Med. 4, 963–966 (1998)
Mu, S., Wang, J., Zhou, G., Peng, W., He, Z., Zhao, Z., Mo, C., Qu, J., Zhang, J.: Transplantation of induced pluripotent stem cells improves functional recovery in Huntington’s disease rat model. PLoS ONE 9, e101185 (2014)
Bachoud-Lévi, A.-C., Rémy, P., Nǵuyen, J.-P., Brugières, P., Lefaucheur, J.-P., Bourdet, C., Baudic, S., Gaura, V., Maison, P., Haddad, B., Boissé, M.-F., Grandmougin, T., Jény, R., Bartolomeo, P., Barba, G.D., Degos, J.-D., Lisovoski, F., Ergis, A.-M., Pailhous, E., Cesaro, P., Hantraye, P., Peschanski, M.: Motor and cognitive improvements in patients with Huntington’s disease after neural transplantation. Lancet 356, 1975–1979 (2000)
Modo, M., Mellodew, K., Cash, D., Fraser, S.E., Meade, T.J., Price, J., Williams, S.C.: Mapping transplanted stem cell migration after a stroke: a serial, in vivo magnetic resonance imaging study. Neuroimage 21, 311–317 (2004)
Guzman, R., Uchida, N., Bliss, T.M., He, D., Christopherson, K.K., Stellwagen, D., Capela, A., Greve, J., Malenka, R.C., Moseley, M.E., Palmer, T.D., Steinberg, G.K.: Long-term monitoring of transplanted human neural stem cells in developmental and pathological contexts with MRI. Proc. Natl. Acad. Sci. U.S.A. 104, 10211–10216 (2007)
Kraitchman, D.L., Gilson, W.D., Lorenz, C.H.: Stem cell therapy: MRI guidance and monitoring. J. Magn. Reson. Imaging 27, 299–310 (2008)
Nicoleau, C., Varela, C., Bonnefond, C., Maury, Y., Bugi, A., Aubry, L., Viegas, P., Bourgois-Rocha, F., Peschanski, M., Perrier, A.L.: Embryonic stem cells neural differentiation qualifies the role of Wnt/β-Catenin signals in human telencephalic specification and regionalization. Stem Cells 31, 1763–1774 (2013)
Dauguet, J., Delzescaux, T., Condé, F., Mangin, J.-F., Ayache, N., Hantraye, P., Frouin, V.: Three-dimensional reconstruction of stained histological slices and 3D non-linear registration with in-vivo MRI for whole baboon brain. J. Neurosci. Methods 164, 191–204 (2007)
Ourselin, S., Roche, A., Prima, S., Ayache, N.: Block matching: a general framework to improve robustness of rigid registration of medical images. In: Delp, S.L., DiGoia, A.M., Jaramaz, B. (eds.) MICCAI 2000. LNCS, vol. 1935, pp. 557–566. Springer, Heidelberg (2000)
Rueckert, D., Sonoda, L.I., Hayes, C., Hill, D.L., Leach, M.O., Hawkes, D.J.: Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans. Med. Imaging 18, 712–721 (1999)
Mattes, D., Haynor, D.R., Vesselle, H., Lewellen, T.K., Eubank, W.: PET-CT image registration in the chest using free-form deformations. IEEE Trans. Med. Imaging 22, 120–128 (2003)
Golland, P., Fischl, B.: Permutation tests for classification: towards statistical significance in image-based studies. Inf. Process. Med. Imaging. 18, 330–341 (2003)
Ojala, M., Garriga, G.C.: Permutation tests for studying classifier performance. In: 2009 Ninth IEEE International Conference on Data Mining, pp. 908–913. IEEE (2009)
Bakker, R., Tiesinga, P., Kötter, R.: The scalable brain atlas: instant web-based access to public brain atlases and related content, pp. 353–366 (2013). arXiv Preprint: arXiv1312.6310
Acknowledgements
This study was partially supported by the French National Agency for Research (ANR-2010-RFCS-003 “HD-SCT”) and by the Laboratoire d’Excellence Revive (Investissement d’Avenir; ANR-10-LABX-73). We thank Martine Guillermier, Susannah Williams, Aurore Bugi and Nicolas Souedet for their contribution to this work.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this paper
Cite this paper
Balbastre, Y. et al. (2016). A Quantitative Approach to Characterize MR Contrasts with Histology. In: Crimi, A., Menze, B., Maier, O., Reyes, M., Handels, H. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2015. Lecture Notes in Computer Science(), vol 9556. Springer, Cham. https://doi.org/10.1007/978-3-319-30858-6_10
Download citation
DOI: https://doi.org/10.1007/978-3-319-30858-6_10
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-30857-9
Online ISBN: 978-3-319-30858-6
eBook Packages: Computer ScienceComputer Science (R0)