Abstract
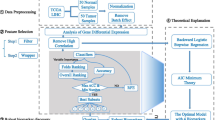
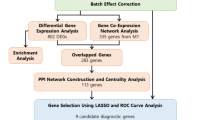
In our previous research we developed a methodology for extracting significant genes that indicate colorectal cancer (CRC). By using those biomarker genes we proposed an intelligent modelling of their gene expression distributions and used them in the Bayes’ theorem in order to achieve highly precise classification of patients in one of the classes carcinogenic, or healthy. The main objective of our new research is to subside the biomarkers set without degrading the sensitivity and specificity of the classifier. We want to eliminate the biomarkers that do not play an important role in the classification process. To achieve this goal, we propose a novel approach for biomarkers detection based on iterative Bayesian classification. The new Leave-one-out method aims to extract the biomarkers essential for the classification process, i.e. if they are left-out, the classification shows remarkably degraded results. Taking into account only the reduced set of biomarkers, we produced an improved version of our Bayesian classifier when classifying new patients. Another advantage of our approach is using the new biomarkers set in the Gene Ontology (GO) analysis in order to get more precise information on the colorectal cancer’s biomarkers’ biological and molecular functions.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Preview
Unable to display preview. Download preview PDF.
Similar content being viewed by others
References
Simjanoska, M., Madevska Bogdanova, A., Popeska, Z.: Bayesian posterior probability classification of colorectal cancer probed with Affymetrix microarray technology. In: 2013 36th International Convention on Information & Communication Technology Electronics & Microelectronics (MIPRO), pp. 959–964. IEEE (2013)
Simjanoska, M., Madevska Bogdanova, A., Panov, S.: Gene ontology analysis of colorectal cancer biomarkers probed with Affymetrix and Illumina microarrays. In: Proceedings of the 5th International Joint Conference on Computational Intelligence, IJCCI 2013, pp. 396–406. IJCCI (2013)
Lascorz, J., Chen, B., Hemminki, K., Försti, A.: Consensus pathways implicated in prognosis of colorectal cancer identified through systematic enrichment analysis of gene expression profiling studies. PloS One 6(4), e18867 (2011)
Xu, Y., Xu, Q., Yang, L., Liu, F., Ye, X., Wu, F., Ni, S., Tan, C., Cai, G., Meng, X., et al.: Gene expression analysis of peripheral blood cells reveals toll-like receptor pathway deregulation in colorectal cancer. PloS One 8(5), e62870 (2013)
Chan, S.K., Griffith, O.L., Tai, I.T., Jones, S.J.: Meta-analysis of colorectal cancer gene expression profiling studies identifies consistently reported candidate biomarkers. Cancer Epidemiology Biomarkers & Prevention 17(3), 543–552 (2008)
Jiang, W., Li, X., Rao, S., Wang, L., Du, L., Li, C., Wu, C., Wang, H., Wang, Y., Yang, B.: Constructing disease-specific gene networks using pair-wise relevance metric: application to colon cancer identifies interleukin 8, desmin and enolase 1 as the central elements. BMC Systems Biology 2(1), 72 (2008)
Wu, Z., Aryee, M.: Subset quantile normalization using negative control features. Journal of Computational Biology 17(10), 1385–1395 (2010)
Needham, C., Manfield, I., Bulpitt, A., Gilmartin, P., Westhead, D.: From gene expression to gene regulatory networks in arabidopsis thaliana. BMC Systems Biology 3(1), 85 (2009)
Hui, Y., Kang, T., Xie, L., Yuan-Yuan, L.: Digout: Viewing differential expression genes as outliers. Journal of Bioinformatics and Computational Biology 8(suppl. 01), 161–175 (2010)
GLOBOCAN (2008), http://globocan.iarc.fr/factsheets/cancers/colorectal.asp
Wang, K., Li, M., Hakonarson, H.: Analysing biological pathways in genome-wide association studies. Nature Reviews Genetics 11(12), 843–854 (2010)
Ashburner, M., Ball, C.A., Blake, J.A., Botstein, D., Butler, H., Cherry, J.M., Davis, A.P., Dolinski, K., Dwight, S.S., Eppig, J.T., et al.: Gene ontology: tool for the unification of biology. Nature Genetics 25(1), 25 (2000)
Harris, M., Clark, J., Ireland, A., Lomax, J., Ashburner, M., Foulger, R., Eilbeck, K., Lewis, S., Marshall, B., Mungall, C., et al.: The gene ontology (go) database and informatics resource. Nucleic Acids Research 32(Database issue), D258 (2004)
Zheng, Q., Wang, X.J.: Goeast: a web-based software toolkit for gene ontology enrichment analysis. Nucleic Acids Research 36(suppl. 2), W358–W363 (2008)
Simjanoska, M., Bogdanova, A.M., Popeska, Z.: Bayesian multiclass classification of gene expression colorectal cancer stages. In: Trajkovik, V., Anastas, M. (eds.) ICT Innovations 2013. AISC, vol. 231, pp. 177–186. Springer, Heidelberg (2013)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this paper
Cite this paper
Simjanoska, M., Madevska Bogdanova, A. (2015). Novel Methodology for CRC Biomarkers Detection with Leave-One-Out Bayesian Classification. In: Bogdanova, A., Gjorgjevikj, D. (eds) ICT Innovations 2014. ICT Innovations 2014. Advances in Intelligent Systems and Computing, vol 311. Springer, Cham. https://doi.org/10.1007/978-3-319-09879-1_23
Download citation
DOI: https://doi.org/10.1007/978-3-319-09879-1_23
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-09878-4
Online ISBN: 978-3-319-09879-1
eBook Packages: EngineeringEngineering (R0)