Abstract
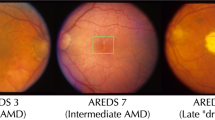
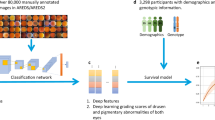
In the elderly, degenerative diseases often develop differently over time for individual patients. For optimal treatment, physicians and patients would like to know how much time is left for them until symptoms reach a certain stage. However, compared to simple disease detection tasks, disease progression modeling has received much less attention. In addition, most existing models are black-box models which provide little insight into the mechanisms driving the prediction. Here, we introduce an interpretable-by-design survival model to predict the progression of age-related macular degeneration (AMD) from fundus images. Our model not only achieves state-of-the-art prediction performance compared to black-box models but also provides a sparse map of local evidence of AMD progression for individual patients. Our evidence map faithfully reflects the decision-making process of the model in contrast to widely used post-hoc saliency methods. Furthermore, we show that the identified regions mostly align with established clinical AMD progression markers. We believe that our method may help to inform treatment decisions and may lead to better insights into imaging biomarkers indicative of disease progression. The project’s code is available at https://github.com/berenslab/interpretable-deep-survival-analysis.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Age-related eye disease study research group: the age-related eye disease study (AREDS): design implications AREDS report No. 1. Controlled Clin. Trials 20(6), 573–600 (1999). https://doi.org/10.1016/S0197-2456(99)00031-8
Age-related eye disease study research group: a randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss: AREDS Report No. 8. Arch. Ophthalmol. 119(10), 1417–1436 (2001). https://doi.org/10.1001/archopht.119.10.1417
Arun, N., et al.: Assessing the trustworthiness of saliency maps for localizing abnormalities in medical imaging. Radiol. Artif. Intell. 3(6), e200267 (2021). https://doi.org/10.1148/ryai.2021200267
Babenko, B., et al.: Predicting progression of age-related macular degeneration from fundus images using deep learning (2019). arXiv:1904.05478 [cs.CV]
Blanche, P., Dartigues, J.F., Jacqmin-Gadda, H.: Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat. Med. 32(30), 5381–5397 (2013). https://doi.org/10.1002/sim.5958
Brendel, W., Bethge, M.: Approximating CNNs with bag-of-local-features models works surprisingly well on ImageNet. In: International Conference on Learning Representations (2019)
Breslow, N.: Discussion of professor cox’s paper. J. Roy. Stat. Soc. B 34, 216–217 (1972)
Cox, D.R.: Regression models and life-tables. J. Roy. Stat. Soc.: Ser. B (Methodol.) 34(2), 187–202 (1972). https://doi.org/10.1111/j.2517-6161.1972.tb00899.x
Dimas, G., Cholopoulou, E., Iakovidis, D.K.: E pluribus unum interpretable convolutional neural networks. Sci. Rep. 13(1), 11421 (2023). https://doi.org/10.1038/s41598-023-38459-1
Djoumessi, K.R., et al.: Sparse activations for interpretable disease grading. In: Medical Imaging with Deep Learning (2023)
Faraggi, D., Simon, R.: A neural network model for survival data. Stat. Med. 14(1), 73–82 (1995). https://doi.org/10.1002/sim.4780140108
Fleckenstein, M., et al.: Age-related macular degeneration. Nat. Rev. Dis. Primers. 7(1), 31 (2021). https://doi.org/10.1038/s41572-021-00265-2
Grassmann, F., et al.: A deep learning algorithm for prediction of age-related eye disease study severity scale for age-related macular degeneration from color fundus photography. Ophthalmology 125(9), 1410–1420 (2018). https://doi.org/10.1016/j.ophtha.2018.02.037
Grote, T.: The allure of simplicity: on interpretable machine learning models in healthcare. Phil. Med. 4(1) (2023). https://doi.org/10.5195/pom.2023.139
Huang, Y., Lin, L., Cheng, P., Lyu, J., Tam, R., Tang, X.: Identifying the key components in ResNet-50 for diabetic retinopathy grading from fundus images: a systematic investigation. Diagnostics 13(10), 1664 (2023). https://doi.org/10.3390/diagnostics13101664
Katzman, J.L., Shaham, U., Cloninger, A., Bates, J., Jiang, T., Kluger, Y.: DeepSurv: personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC Med. Res. Methodol. 18(1), 24 (2018). https://doi.org/10.1186/s12874-018-0482-1
Lambert, J., Chevret, S.: Summary measure of discrimination in survival models based on cumulative/dynamic time-dependent roc curves. Stat. Methods Med. Res. 25(5), 2088–2102 (2016)
Nagpal, C., Potosnak, W., Dubrawski, A.: auton-survival: an open-source package for regression, counterfactual estimation, evaluation and phenotyping with censored time-to-event data (2022). https://doi.org/10.48550/arXiv.2204.07276. arXiv:2204.07276 [cs, stat]
Peng, Y., et al.: DeepSeeNet: a deep learning model for automated classification of patient-based age-related macular degeneration severity from color fundus photographs. Ophthalmology 126(4), 565–575 (2019). https://doi.org/10.1016/j.ophtha.2018.11.015
Pölsterl, S.: scikit-survival: a library for time-to-event analysis built on top of scikit-learn. J. Mach. Learn. Res. 21(212), 1–6 (2020)
Rudin, C.: Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat. Mach. Intell. 1(5), 206–215 (2019). https://doi.org/10.1038/s42256-019-0048-x
Saporta, A., et al.: Benchmarking saliency methods for chest X-ray interpretation. Nat. Mach. Intell. 4(10), 867–878 (2022). https://doi.org/10.1038/s42256-022-00536-x
Wiegrebe, S., Kopper, P., Sonabend, R., Bischl, B., Bender, A.: Deep learning for survival analysis: a review. Artif. Intell. Rev. 57(3), 65 (2024). https://doi.org/10.1007/s10462-023-10681-3
Xu-Darme, R., Quénot, G., Chihani, Z., Rousset, M.C.: Sanity checks and improvements for patch visualisation in prototype-based image classification (2023). https://doi.org/10.48550/arXiv.2302.08508. arXiv:2302.08508 [cs]
Yan, Q., et al.: Deep-learning-based prediction of late age-related macular degeneration progression. Nat. Mach. Intell. 2(2), 141–150 (2020). https://doi.org/10.1038/s42256-020-0154-9
Yin, C., Moroi, S.E., Zhang, P.: Predicting age-related macular degeneration progression with contrastive attention and time-aware LSTM. In: KDD: proceedings. International Conference on Knowledge Discovery & Data Mining, vol. 2022, pp. 4402–4412 (2022). https://doi.org/10.1145/3534678.3539163
Acknowledgments
This project was supported by the Hertie Foundation, the Else Kröner Medical Scientist Kolleg “ClinbrAIn: Artificial Intelligence for Clinical Brain Research”, the Bundesministerium für Bildung und Forschung through the EYERISK project (BMBF, FKZ 01ZZ2319A) and the German Science Foundation (BE5601/8-1 and the Excellence Cluster 2064 “Machine Learning—New Perspectives for Science”, project number 390727645). The authors thank the AREDS participants, and the AREDS Research Group for their valuable contribution to this research. Funding support for AREDS was provided by the National Eye Institute (N01-EY-0-2127).
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Ethics declarations
Disclosure of Interests
The authors declare no relevant competing interests.
1 Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Gervelmeyer, J. et al. (2024). Interpretable-by-Design Deep Survival Analysis for Disease Progression Modeling. In: Linguraru, M.G., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2024. MICCAI 2024. Lecture Notes in Computer Science, vol 15010. Springer, Cham. https://doi.org/10.1007/978-3-031-72117-5_47
Download citation
DOI: https://doi.org/10.1007/978-3-031-72117-5_47
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-72116-8
Online ISBN: 978-3-031-72117-5
eBook Packages: Computer ScienceComputer Science (R0)