Abstract
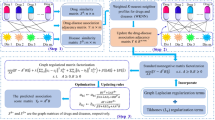
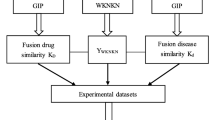
Matrix factorization (MF) has been widely used in drug discovery for link prediction, which aims to reveal new drug-target links by integrating drug-drug and target-target similarity information with a drug-target interaction matrix. The MF method is based on the assumption that similar drugs share similar targets and vice versa. However, one major disadvantage is that only one similarity metric is used in MF models, which is not enough to represent the similarity between drugs or targets. In this work, we develop a similarity fusion enhanced MF model to incorporate different types of similarity for novel drug-target link prediction. We apply the proposed model on a drug-virus association dataset for anti-COVID drug prioritization, and compare the performance with other existing MF models developed for COVID. The results show that the similarity fusion method can provide more useful information for drug-drug and virus-virus similarity and hence improve the performance of MF models. The top 10 drugs as prioritized by our model are provided, together with supporting evidence from literature.
Y. Li—Supported by China Scholarship Council.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Who coronavirus (covid-19) dashboard. https://covid19.who.int/
Ahlgren, N.A., Ren, J., Lu, Y.Y., Fuhrman, J.A., Sun, F.: Alignment-free oligonucleotide frequency dissimilarity measure improves prediction of hosts from metagenomically-derived viral sequences. Nucleic Acids Res. 45(1), 39–53 (2017)
Aiyegbusi, O.L., et al.: Symptoms, complications and management of long covid: a review. J. R. Soc. Med. 114(9), 428–442 (2021)
Basu, D., Chavda, V.P., Mehta, A.A.: Therapeutics for covid-19 and post covid-19 complications: an update. Current Res. Pharmacol. Drug Discovery, 100086 (2022)
Björnsson, H., Venegas, S.: A manual for EOF and SVD analyses of climatic data. CCGCR Report 97(1), 112–134 (1997)
Chen, R., Liu, X., Jin, S., Lin, J., Liu, J.: Machine learning for drug-target interaction prediction. Molecules 23(9), 2208 (2018)
Ciotti, M., Ciccozzi, M., Terrinoni, A., Jiang, W.C., Wang, C.B., Bernardini, S.: The covid-19 pandemic. Crit. Rev. Clin. Lab. Sci. 57(6), 365–388 (2020)
Dolgin, E.: Omicron is supercharging the covid vaccine booster debate. Nature 10 (2021)
Elmorsy, M.A., El-Baz, A.M., Mohamed, N.H., Almeer, R., Abdel-Daim, M.M., Yahya, G.: In silico screening of potent inhibitors against covid-19 key targets from a library of FDA-approved drugs. Environ. Sci. Pollut. Res. 29(8), 12336–12346 (2022)
Ezzat, A., Zhao, P., Wu, M., Li, X.L., Kwoh, C.K.: Drug-target interaction prediction with graph regularized matrix factorization. IEEE/ACM Trans. Comput. Biol. Bioinf. 14(3), 646–656 (2016)
Gottlieb, A., Stein, G.Y., Ruppin, E., Sharan, R.: Predict: a method for inferring novel drug indications with application to personalized medicine. Mol. Syst. Biol. 7(1), 496 (2011)
Griffith, M., et al.: Dgidb: mining the druggable genome. Nat. Methods 10(12), 1209–1210 (2013)
Hanley, J.A., McNeil, B.J.: The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143(1), 29–36 (1982)
Hattori, M., Tanaka, N., Kanehisa, M., Goto, S.: Simcomp/subcomp: chemical structure search servers for network analyses. Nucleic Acids Res. 38(Suppl-2), W652–W656 (2010)
Hizukuri, Y., Sawada, R., Yamanishi, Y.: Predicting target proteins for drug candidate compounds based on drug-induced gene expression data in a chemical structure-independent manner. BMC Med. Genomics 8(1), 1–10 (2015)
Huang, L., Luo, H., Li, S., Wu, F.X., Wang, J.: Drug-drug similarity measure and its applications. Briefings Bioinform. 22(4), bbaa265 (2021)
Kanehisa, M., et al.: KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36(suppl-1), 480–484 (2007)
Koren, Y., Bell, R., Volinsky, C.: Matrix factorization techniques for recommender systems. Computer 42(8), 30–37 (2009)
Kováč, I.M.J.Č.G., Hudecová, M.P.L.: Triazavirin might be the new hope to fight severe acute respiratory syndrome coronavirus 2 (sars-cov-2). Ceska a Slovenska farmacie: casopis Ceske farmaceuticke spolecnosti a Slovenske farmaceuticke spolecnosti 70(1), 18–25 (2021)
Liu, H., Sun, J., Guan, J., Zheng, J., Zhou, S.: Improving compound-protein interaction prediction by building up highly credible negative samples. Bioinformatics 31(12), i221–i229 (2015)
Manabe, T., Kambayashi, D., Akatsu, H., Kudo, K.: Favipiravir for the treatment of patients with covid-19: a systematic review and meta-analysis. BMC Infect. Dis. 21(1), 1–13 (2021)
Meng, Y., Jin, M., Tang, X., Xu, J.: Drug repositioning based on similarity constrained probabilistic matrix factorization: covid-19 as a case study. Appl. Soft Comput. 103, 107135 (2021)
Mohamed, K., Yazdanpanah, N., Saghazadeh, A., Rezaei, N.: Computational drug discovery and repurposing for the treatment of covid-19: a systematic review. Bioorg. Chem. 106, 104490 (2021)
Mongia, A., Jain, S., Chouzenoux, E., Majumdar, A.: Deepvir: graphical deep matrix factorization for in silico antiviral repositioning-application to covid-19. J. Comput. Biol. (2022)
Mongia, A., Saha, S.K., Chouzenoux, E., Majumdar, A.: A computational approach to aid clinicians in selecting anti-viral drugs for covid-19 trials. Sci. Rep. 11(1), 1–12 (2021)
Muratov, E.N., et al.: A critical overview of computational approaches employed for covid-19 drug discovery. Chem. Soc. Rev. (2021)
Park, K., Kim, D., Ha, S., Lee, D.: Predicting pharmacodynamic drug-drug interactions through signaling propagation interference on protein-protein interaction networks. PLoS ONE 10(10), e0140816 (2015)
Ramachandran, R., et al.: Phase iii, randomized, double-blind, placebo controlled trial of efficacy, safety and tolerability of antiviral drug umifenovir vs standard care of therapy in non-severe covid-19 patients. Int. J. Infect. Dis. 115, 62–69 (2022)
Rogers, D., Hahn, M.: Extended-connectivity fingerprints. J. Chem. Inf. Model. 50(5), 742–754 (2010)
Tang, X., Cai, L., Meng, Y., Xu, J., Lu, C., Yang, J.: Indicator regularized non-negative matrix factorization method-based drug repurposing for covid-19. Front. Immunol. 11, 3824 (2021)
Tanimoto, T.T.: Elementary mathematical theory of classification and prediction (1958)
Vilar, S., Hripcsak, G.: The role of drug profiles as similarity metrics: applications to repurposing, adverse effects detection and drug-drug interactions. Brief. Bioinform. 18(4), 670–681 (2017)
Zhang, C.x., et al.: Peramivir, an anti-influenza virus drug, exhibits potential anti-cytokine storm effects. bioRxiv (2020)
Zhang, W., Chen, Y., Li, D., Yue, X.: Manifold regularized matrix factorization for drug-drug interaction prediction. J. Biomed. Inform. 88, 90–97 (2018)
Zhang, W., Liu, X., Chen, Y., Wu, W., Wang, W., Li, X.: Feature-derived graph regularized matrix factorization for predicting drug side effects. Neurocomputing 287, 154–162 (2018)
Zhang, W., et al.: Predicting drug-disease associations by using similarity constrained matrix factorization. BMC Bioinform. 19(1), 1–12 (2018)
Zhang, W., Zou, H., Luo, L., Liu, Q., Wu, W., Xiao, W.: Predicting potential side effects of drugs by recommender methods and ensemble learning. Neurocomputing 173, 979–987 (2016)
Zhang, Z.C., Zhang, X.F., Wu, M., Ou-Yang, L., Zhao, X.M., Li, X.L.: A graph regularized generalized matrix factorization model for predicting links in biomedical bipartite networks. Bioinformatics 36(11), 3474–3481 (2020)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Li, Y., Xu, X., Tsoka, S. (2023). A Matrix Factorization-Based Drug-Virus Link Prediction Method for SARS-CoV-2 Drug Prioritization. In: Nicosia, G., et al. Machine Learning, Optimization, and Data Science. LOD 2022. Lecture Notes in Computer Science, vol 13810. Springer, Cham. https://doi.org/10.1007/978-3-031-25599-1_4
Download citation
DOI: https://doi.org/10.1007/978-3-031-25599-1_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-25598-4
Online ISBN: 978-3-031-25599-1
eBook Packages: Computer ScienceComputer Science (R0)