Abstract
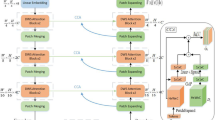
In the past few years, convolutional neural networks (CNNs) have achieved milestones in medical image analysis. In particular, deep neural networks based on U-shaped architecture and skip-connections have been widely applied in various medical image tasks. However, although CNN has achieved excellent performance, it cannot learn global semantic information interaction well due to the locality of convolution operation. In this paper, we propose Swin-Unet, which is an Unet-like pure Transformer for medical image segmentation. The tokenized image patches are fed into the Transformer-based U-shaped Encoder-Decoder architecture with skip-connections for local-global semantic feature learning. Specifically, we use a hierarchical Swin Transformer with shifted windows as the encoder to extract context features. And a symmetric Swin Transformer-based decoder with a patch expanding layer is designed to perform the up-sampling operation to restore the spatial resolution of the feature maps. Under the direct down-sampling and up-sampling of the inputs and outputs by \(4{\times }\), experiments on multi-organ and cardiac segmentation tasks demonstrate that the pure Transformer-based U-shaped Encoder-Decoder network outperforms those methods with full-convolution or the combination of transformer and convolution. The codes have been publicly available at the link (https://github.com/HuCaoFighting/Swin-Unet).
H. Cao and Y. Wang—Work done as an intern in Huawei Technologies.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Carion, N., Massa, F., Synnaeve, G., Usunier, N., Kirillov, A., Zagoruyko, S.: End-to-end object detection with transformers. In: Vedaldi, A., Bischof, H., Brox, T., Frahm, J.-M. (eds.) ECCV 2020. LNCS, vol. 12346, pp. 213–229. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-58452-8_13
Chen, J., et al.: TransUNet: transformers make strong encoders for medical image segmentation. CoRR abs/2102.04306 (2021)
Chen, L.C., Papandreou, G., Kokkinos, I., Murphy, K., Yuille, A.L.: DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 40(4), 834–848 (2018). https://doi.org/10.1109/TPAMI.2017.2699184
Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T., Ronneberger, O.: 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 424–432. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_49
Devlin, J., Chang, M.W., Lee, K., Toutanova, K.: BERT: pre-training of deep bidirectional transformers for language understanding. In: Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers), pp. 4171–4186. Association for Computational Linguistics, Minneapolis, June 2019. https://doi.org/10.18653/v1/N19-1423. https://www.aclweb.org/anthology/N19-1423
Dosovitskiy, A., et al.: An image is worth \(16\times 16\) words: transformers for image recognition at scale. In: International Conference on Learning Representations (2021)
Isensee, F., Jaeger, P.F., Kohl, S.A., Petersen, J., Maier-Hein, K.H.: nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18(2), 203–211 (2021)
Fu, S., et al.: Domain adaptive relational reasoning for 3D multi-organ segmentation. In: Martel, A.L., et al. (eds.) MICCAI 2020. LNCS, vol. 12261, pp. 656–666. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-59710-8_64
Gu, Z., et al.: CE-Net: context encoder network for 2D medical image segmentation. IEEE Trans. Med. Imaging 38(10), 2281–2292 (2019). https://doi.org/10.1109/TMI.2019.2903562
Han, K., Xiao, A., Wu, E., Guo, J., Xu, C., Wang, Y.: Transformer in transformer. In: Ranzato, M., Beygelzimer, A., Dauphin, Y., Liang, P., Vaughan, J.W. (eds.) Advances in Neural Information Processing Systems, vol. 34, pp. 15908–15919. Curran Associates, Inc. (2021). https://proceedings.neurips.cc/paper/2021/file/854d9fca60b4bd07f9bb215d59ef5561-Paper.pdf
Hatamizadeh, A., et al.: UNETR: transformers for 3D medical image segmentation. In: 2022 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), pp. 1748–1758 (2022). https://doi.org/10.1109/WACV51458.2022.00181
Held, K., Kops, E., Krause, B., Wells, W., Kikinis, R., Muller-Gartner, H.W.: Markov random field segmentation of brain MR images. IEEE Trans. Med. Imaging 16(6), 878–886 (1997). https://doi.org/10.1109/42.650883
Hu, H., Gu, J., Zhang, Z., Dai, J., Wei, Y.: Relation networks for object detection. In: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 3588–3597 (2018). https://doi.org/10.1109/CVPR.2018.00378
Hu, H., Zhang, Z., Xie, Z., Lin, S.: Local relation networks for image recognition. In: 2019 IEEE/CVF International Conference on Computer Vision (ICCV), pp. 3463–3472 (2019). https://doi.org/10.1109/ICCV.2019.00356
Huang, H., et al.: UNet 3+: a full-scale connected UNet for medical image segmentation (2020)
Jin, Q., Meng, Z., Sun, C., Cui, H., Su, R.: RA-UNet: a hybrid deep attention-aware network to extract liver and tumor in CT scans. Front. Bioeng. Biotechnol. 8, 1471 (2020)
Li, X., Chen, H., Qi, X., Dou, Q., Fu, C.W., Heng, P.A.: H-DenseUNet: hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans. Med. Imaging 37(12), 2663–2674 (2018). https://doi.org/10.1109/TMI.2018.2845918
Liu, Z., et al.: Swin transformer: hierarchical vision transformer using shifted windows. In: 2021 IEEE/CVF International Conference on Computer Vision (ICCV), pp. 9992–10002 (2021). https://doi.org/10.1109/ICCV48922.2021.00986
Milletari, F., Navab, N., Ahmadi, S.A.: V-Net: fully convolutional neural networks for volumetric medical image segmentation. In: 2016 Fourth International Conference on 3D Vision (3DV), pp. 565–571 (2016). https://doi.org/10.1109/3DV.2016.79
Oktay, O., et al.: Attention U-Net: learning where to look for the pancreas. In: IMIDL Conference (2018)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Schlemper, J., et al.: Attention gated networks: learning to leverage salient regions in medical images. Med. Image Anal. 53, 197–207 (2019). https://doi.org/10.1016/j.media.2019.01.012
Touvron, H., Cord, M., Douze, M., Massa, F., Sablayrolles, A., Jegou, H.: Training data-efficient image transformers distillation through attention. In: Meila, M., Zhang, T. (eds.) Proceedings of the 38th International Conference on Machine Learning. Proceedings of Machine Learning Research, vol. 139, pp. 10347–10357. PMLR, 18–24 July 2021. https://proceedings.mlr.press/v139/touvron21a.html
Touvron, H., Cord, M., Sablayrolles, A., Synnaeve, G., Jégou, H.: Going deeper with image transformers. In: 2021 IEEE/CVF International Conference on Computer Vision (ICCV), pp. 32–42 (2021). https://doi.org/10.1109/ICCV48922.2021.00010
Tsai, A., et al.: A shape-based approach to the segmentation of medical imagery using level sets. IEEE Trans. Med. Imaging 22(2), 137–154 (2003). https://doi.org/10.1109/TMI.2002.808355
Valanarasu, J.M.J., Oza, P., Hacihaliloglu, I., Patel, V.M.: Medical transformer: gated axial-attention for medical image segmentation. In: de Bruijne, M., et al. (eds.) MICCAI 2021. LNCS, vol. 12901, pp. 36–46. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-87193-2_4
Vaswani, A., et al.: Attention is all you need. In: Advances in Neural Information Processing Systems, vol. 30. Curran Associates, Inc. (2017)
Wang, W., et al.: Pyramid vision transformer: a versatile backbone for dense prediction without convolutions. In: 2021 IEEE/CVF International Conference on Computer Vision (ICCV), pp. 548–558 (2021). https://doi.org/10.1109/ICCV48922.2021.00061
Wang, W., Chen, C., Ding, M., Yu, H., Zha, S., Li, J.: TransBTS: multimodal brain tumor segmentation using transformer. In: de Bruijne, M., et al. (eds.) MICCAI 2021. LNCS, vol. 12901, pp. 109–119. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-87193-2_11
Wang, X., Girshick, R., Gupta, A., He, K.: Non-local neural networks. In: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 7794–7803 (2018). https://doi.org/10.1109/CVPR.2018.00813
Xiao, X., Lian, S., Luo, Z., Li, S.: Weighted Res-UNet for high-quality retina vessel segmentation. In: 2018 9th International Conference on Information Technology in Medicine and Education (ITME), pp. 327–331 (2018)
Xie, Y., Zhang, J., Shen, C., Xia, Y.: CoTr: efficiently bridging CNN and transformer for 3D medical image segmentation. In: de Bruijne, M., et al. (eds.) MICCAI 2021. LNCS, vol. 12903, pp. 171–180. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-87199-4_16
Zhang, Y., Liu, H., Hu, Q.: TransFuse: fusing transformers and CNNs for medical image segmentation. In: de Bruijne, M., et al. (eds.) MICCAI 2021. LNCS, vol. 12901, pp. 14–24. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-87193-2_2
Zhao, H., Shi, J., Qi, X., Wang, X., Jia, J.: Pyramid scene parsing network. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 6230–6239 (2017). https://doi.org/10.1109/CVPR.2017.660
Zhou, Z., Rahman Siddiquee, M.M., Tajbakhsh, N., Liang, J.: UNet++: a nested U-Net architecture for medical image segmentation. In: Stoyanov, D., et al. (eds.) DLMIA/ML-CDS -2018. LNCS, vol. 11045, pp. 3–11. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00889-5_1
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Cao, H. et al. (2023). Swin-Unet: Unet-Like Pure Transformer for Medical Image Segmentation. In: Karlinsky, L., Michaeli, T., Nishino, K. (eds) Computer Vision – ECCV 2022 Workshops. ECCV 2022. Lecture Notes in Computer Science, vol 13803. Springer, Cham. https://doi.org/10.1007/978-3-031-25066-8_9
Download citation
DOI: https://doi.org/10.1007/978-3-031-25066-8_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-25065-1
Online ISBN: 978-3-031-25066-8
eBook Packages: Computer ScienceComputer Science (R0)