Compositional differences within and between eukaryotic genomes
- PMID: 9294192
- PMCID: PMC23344
- DOI: 10.1073/pnas.94.19.10227
Compositional differences within and between eukaryotic genomes
Abstract
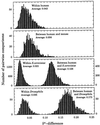
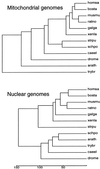
Eukaryotic genome similarity relationships are inferred using sequence information derived from large aggregates of genomic sequences. Comparisons within and between species sample sequences are based on the profile of dinucleotide relative abundance values (The profile is rho*XY = f*XY/f*Xf*Y for all XY, where f*X denotes the frequency of the nucleotide X and f*XY denotes the frequency of the dinucleotide XY, both computed from the sequence concatenated with its inverted complement). Previous studies with respect to prokaryotes and this study document that profiles of different DNA sequence samples (sample size >/=50 kb) from the same organism are generally much more similar to each other than they are to profiles from other organisms, and that closely related organisms generally have more similar profiles than do distantly related organisms. On this basis we refer to the collection (rho*XY) as the genome signature. This paper identifies rho*XY extremes and compares genome signature differences for a diverse range of eukaryotic species. Interpretations on the mechanisms maintaining these profile differences center on genome-wide replication, repair, DNA structures, and context-dependent mutational biases. It is also observed that mitochondrial genome signature differences between species parallel the corresponding nuclear genome signature differences despite large differences between corresponding mitochondrial and nuclear signatures. The genome signature differences also have implications for contrasts between rodents and other mammals, and between monocot and dicot plants, as well as providing evidence for similarities among fungi and the diversity of protists.
Figures
Similar articles
-
Compositional biases of bacterial genomes and evolutionary implications.J Bacteriol. 1997 Jun;179(12):3899-913. doi: 10.1128/jb.179.12.3899-3913.1997. J Bacteriol. 1997. PMID: 9190805 Free PMC article.
-
Genome signature comparisons among prokaryote, plasmid, and mitochondrial DNA.Proc Natl Acad Sci U S A. 1999 Aug 3;96(16):9184-9. doi: 10.1073/pnas.96.16.9184. Proc Natl Acad Sci U S A. 1999. PMID: 10430917 Free PMC article.
-
Additive methods for genomic signatures.BMC Bioinformatics. 2016 Aug 22;17(1):313. doi: 10.1186/s12859-016-1157-8. BMC Bioinformatics. 2016. PMID: 27549194 Free PMC article.
-
Dinucleotide relative abundance extremes: a genomic signature.Trends Genet. 1995 Jul;11(7):283-90. doi: 10.1016/s0168-9525(00)89076-9. Trends Genet. 1995. PMID: 7482779 Review.
-
Global dinucleotide signatures and analysis of genomic heterogeneity.Curr Opin Microbiol. 1998 Oct;1(5):598-610. doi: 10.1016/s1369-5274(98)80095-7. Curr Opin Microbiol. 1998. PMID: 10066522 Review.
Cited by
-
Dual Roles of Host Zinc Finger Proteins in Viral RNA Regulation: Decay or Stabilization.Int J Mol Sci. 2024 Oct 17;25(20):11138. doi: 10.3390/ijms252011138. Int J Mol Sci. 2024. PMID: 39456919 Free PMC article. Review.
-
Codon Usage Analysis Reveals Distinct Evolutionary Patterns and Host Adaptation Strategies in Duck Hepatitis Virus 1 (DHV-1) Phylogroups.Viruses. 2024 Aug 29;16(9):1380. doi: 10.3390/v16091380. Viruses. 2024. PMID: 39339856 Free PMC article.
-
Depletion of CpG dinucleotides in bacterial genomes may represent an adaptation to high temperatures.NAR Genom Bioinform. 2024 Jul 27;6(3):lqae088. doi: 10.1093/nargab/lqae088. eCollection 2024 Sep. NAR Genom Bioinform. 2024. PMID: 39071851 Free PMC article.
-
Molecular Characterization and Genome Mechanical Features of Two Newly Isolated Polyvalent Bacteriophages Infecting Pseudomonas syringae pv. garcae.Genes (Basel). 2024 Jan 18;15(1):113. doi: 10.3390/genes15010113. Genes (Basel). 2024. PMID: 38255005 Free PMC article.
-
An Evolutionary Perspective of Codon Usage Pattern, Dinucleotide Composition and Codon Pair Bias in Prunus Necrotic Ringspot Virus.Genes (Basel). 2023 Aug 28;14(9):1712. doi: 10.3390/genes14091712. Genes (Basel). 2023. PMID: 37761852 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases


