SmProt: A Reliable Repository with Comprehensive Annotation of Small Proteins Identified from Ribosome Profiling
- PMID: 34536568
- PMCID: PMC9039559
- DOI: 10.1016/j.gpb.2021.09.002
SmProt: A Reliable Repository with Comprehensive Annotation of Small Proteins Identified from Ribosome Profiling
Abstract
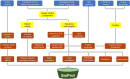
Small proteins specifically refer to proteins consisting of less than 100 amino acids translated from small open reading frames (sORFs), which were usually missed in previous genome annotation. The significance of small proteins has been revealed in current years, along with the discovery of their diverse functions. However, systematic annotation of small proteins is still insufficient. SmProt was specially developed to provide valuable information on small proteins for scientific community. Here we present the update of SmProt, which emphasizes reliability of translated sORFs, genetic variants in translated sORFs, disease-specific sORF translation events or sequences, and remarkably increased data volume. More components such as non-ATG translation initiation, function, and new sources are also included. SmProt incorporated 638,958 unique small proteins curated from 3,165,229 primary records, which were computationally predicted from 419 ribosome profiling (Ribo-seq) datasets or collected from literature and other sources from 370 cell lines or tissues in 8 species (Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, Danio rerio, Saccharomyces cerevisiae, Caenorhabditis elegans, and Escherichia coli). In addition, small protein families identified from human microbiomes were also collected. All datasets in SmProt are free to access, and available for browse, search, and bulk downloads at http://bigdata.ibp.ac.cn/SmProt/.
Keywords: Disease; Ribosome profiling; Small open reading frame; Upstream open reading frame; Variants.
Copyright © 2021 The Authors. Published by Elsevier B.V. All rights reserved.
Figures


Similar articles
-
An update on sORFs.org: a repository of small ORFs identified by ribosome profiling.Nucleic Acids Res. 2018 Jan 4;46(D1):D497-D502. doi: 10.1093/nar/gkx1130. Nucleic Acids Res. 2018. PMID: 29140531 Free PMC article.
-
SmProt: a database of small proteins encoded by annotated coding and non-coding RNA loci.Brief Bioinform. 2018 Jul 20;19(4):636-643. doi: 10.1093/bib/bbx005. Brief Bioinform. 2018. PMID: 28137767
-
Discovery of Unannotated Small Open Reading Frames in Streptococcus pneumoniae D39 Involved in Quorum Sensing and Virulence Using Ribosome Profiling.mBio. 2022 Aug 30;13(4):e0124722. doi: 10.1128/mbio.01247-22. Epub 2022 Jul 19. mBio. 2022. PMID: 35852327 Free PMC article.
-
Evolution of new proteins from translated sORFs in long non-coding RNAs.Exp Cell Res. 2020 Jun 1;391(1):111940. doi: 10.1016/j.yexcr.2020.111940. Epub 2020 Mar 7. Exp Cell Res. 2020. PMID: 32156600 Review.
-
Short open reading frames (sORFs) and microproteins: an update on their identification and validation measures.J Biomed Sci. 2022 Mar 17;29(1):19. doi: 10.1186/s12929-022-00802-5. J Biomed Sci. 2022. PMID: 35300685 Free PMC article. Review.
Cited by
-
Long non-coding RNA-encoded micropeptides: functions, mechanisms and implications.Cell Death Discov. 2024 Oct 23;10(1):450. doi: 10.1038/s41420-024-02175-0. Cell Death Discov. 2024. PMID: 39443468 Free PMC article. Review.
-
Small ORFs, Big Insights: Drosophila as a Model to Unraveling Microprotein Functions.Cells. 2024 Oct 2;13(19):1645. doi: 10.3390/cells13191645. Cells. 2024. PMID: 39404408 Free PMC article. Review.
-
A catalog of small proteins from the global microbiome.Nat Commun. 2024 Aug 31;15(1):7563. doi: 10.1038/s41467-024-51894-6. Nat Commun. 2024. PMID: 39214983 Free PMC article.
-
D-sORF: Accurate Ab Initio Classification of Experimentally Detected Small Open Reading Frames (sORFs) Associated with Translational Machinery.Biology (Basel). 2024 Jul 26;13(8):563. doi: 10.3390/biology13080563. Biology (Basel). 2024. PMID: 39194501 Free PMC article.
-
Peptide-specific chemical language model successfully predicts membrane diffusion of cyclic peptides.bioRxiv [Preprint]. 2024 Aug 9:2024.08.09.607221. doi: 10.1101/2024.08.09.607221. bioRxiv. 2024. PMID: 39149303 Free PMC article. Preprint.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases


